













| General Information: |
|
| Name(s) found: |
cut15 /
SPCC962.03c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 542 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear envelope
[IDA nucleus [IDA cytoplasm [ISS nuclear pore [IEA] |
| Biological Process: |
M phase of mitotic cell cycle
[IMP protein import into nucleus [IDA NLS-bearing substrate import into nucleus [TAS chromosome condensation [IMP cellular response to stress [IGI regulation of mitotic cell cycle [IMP |
| Molecular Function: |
GTP binding
[NAS protein transporter activity [ISS protein binding, bridging [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSASSRFIPE HRRQNYKGKG TFQADELRRR RETQQIEIRK QKREENLNKR RNLVDVQEPA 60
61 EETIPLEQDK ENDLELELQL PDLLKALYSD DIEAQIQATA KFRKALSKET NPPIQKVIDA 120
121 GVVPRFVEFL SHENNLLKFE ASWALTNVAS GSSNQTHVVV EANAVPVFVS LLSSSEQDVR 180
181 EQAVWALGNI AGDSPMCRDH VLQCGVLEPL LNIIESNRRL SMLRNSTWTL SNMCRGKNPQ 240
241 PDWNSISQVI PVLSKLIYTL DEDVLVDALW AISYLSDGAN EKIQAIIDAG IPRRLVELLM 300
301 HPSAQVQTPA LRSVGNIVTG DDVQTQVIIN CGALSALLSL LSSPRDGVRK EACWTISNIT 360
361 AGNSSQIQYV IEANIIPPLI HLLTTADFKI QKEACWAISN ATSGGARRPD QIRYLVEQGA 420
421 IKPLCNLLAC QDNKIIQVAL DGIENILRVG ELDRANNPDK INLYAVYVED AGGMDLIHEC 480
481 QNSSNSEIYQ KAYNIIEKFF GEEDEIEELE PETVGDTFTF GTTQEPAGDF QFSATNAEDM 540
541 AM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
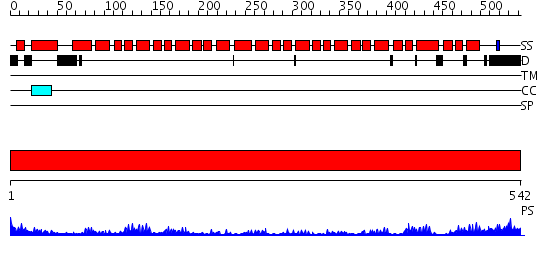
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..542] | 91.154902 | Crystal structure of the exportin Cse1p complexed with its cargo ( Kap60p) and RanGTP |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)