













| General Information: |
|
| Name(s) found: |
ago1 /
SPCC736.11
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 834 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mating-type region heterochromatin
[IDA nuclear telomeric heterochromatin [IDA RITS complex [IDA cytosol [IDA ARC complex [IDA centromeric heterochromatin [IDA |
| Biological Process: |
chromatin silencing at centromere
[IMP regulation of histone H3-K9 methylation [IMP chromosome segregation [IGI regulation of chromatin silencing at centromere [IMP conversion of ds siRNA to ss siRNA involved in RNA interference [IMP [NOT]chromatin silencing at silent mating-type cassette [IMP RNA interference [TAS [NOT]chromatin silencing at telomere [IMP chromatin silencing by small RNA [IMP |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSYKPSSEIA LRPGYGGLGK QITLKANFFQ IISLPNETIN QYHVIVGDGS RVPRKQSQLI 60
61 WNSKEVKQYF GSSWMNSVYD GRSMCWSKGD IADGTIKVNI GSESHPREIE FSIQKSSKIN 120
121 LHTLSQFVNS KYSSDPQVLS SIMFLDLLLK KKPSETLFGF MHSFFTGENG VSLGGGVEAW 180
181 KGFYQSIRPN QGFMSVNVDI SSSAFWRNDS LLQILMEYTD CSNVRDLTRF DLKRLSRKFR 240
241 FLKVTCQHRN NVGTDLANRV YSIEGFSSKS ASDSFFVRRL NGEEQKISVA EYFLENHNVR 300
301 LQYPNLPCIL VKNGAMLPIE FCFVVKGQRY TAKLNSDQTA NMIRFAVQRP FERVQQIDDF 360
361 VHQMDWDTDP YLTQYGMKIQ KKMLEVPARV LETPSIRYGG DCIERPVSGR WNLRGKRFLD 420
421 PPRAPIRSWA VMCFTSTRRL PMRGIENFLQ TYVQTLTSLG INFVMKKPPV LYADIRGSVE 480
481 ELCITLYKKA EQVGNAPPDY LFFILDKNSP EPYGSIKRVC NTMLGVPSQC AISKHILQSK 540
541 PQYCANLGMK INVKVGGINC SLIPKSNPLG NVPTLILGGD VYHPGVGATG VSIASIVASV 600
601 DLNGCKYTAV SRSQPRHQEV IEGMKDIVVY LLQGFRAMTK QQPQRIIYFR DGTSEGQFLS 660
661 VINDELSQIK EACHSLSPKY NPKILVCTTQ KRHHARFFIK NKSDGDRNGN PLPGTIIEKH 720
721 VTHPYQYDFY LISHPSLQGV SVPVHYTVLH DEIQMPPDQF QTLCYNLCYV YARATSAVSL 780
781 VPPVYYAHLV SNLARYQDVT ADDTFVETSE ASMDQEVKPL LALSSKLKTK MWYM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
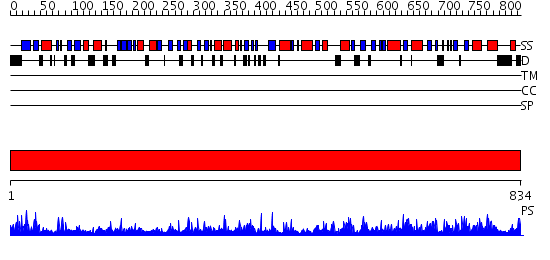
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..834] | 55.39794 | Crystal structure of A. aeolicus Argonaute |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)