













| General Information: |
|
| Name(s) found: |
hhp2 /
SPAC23C4.12
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 400 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS monopolin complex [ISS mitochondrion [IDA |
| Biological Process: |
attachment of spindle microtubules to kinetochore involved in homologous chromosome segregation
[ISS signal transduction [RCA regulation of DNA repair [IMP homologous chromosome segregation [IGI protein amino acid phosphorylation [ISS |
| Molecular Function: |
ATP binding
[ISS protein serine/threonine kinase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVVDIKIGN KYRIGRKIGS GSFGQIYLGL NTVNGEQVAV KLEPLKARHH QLEYEFRVYN 60
61 ILKGNIGIPT IRWFGVTNSY NAMVMDLLGP SLEDLFCYCG RKFTLKTVLL LADQLISRIE 120
121 YVHSKSFLHR DIKPDNFLMK KHSNVVTMID FGLAKKYRDF KTHVHIPYRD NKNLTGTARY 180
181 ASINTHIGIE QSRRDDLESL GYVLLYFCRG SLPWQGLQAD TKEQKYQRIR DTKIGTPLEV 240
241 LCKGLPEEFI TYMCYTRQLS FTEKPNYAYL RKLFRDLLIR KGYQYDYVFD WMILKYQKRA 300
301 AAAAAASATA PPQVTSPMVS QTQPVNPITP NYSSIPLPAE RNPKTPQSFS TNIVQCASPS 360
361 PLPLSFRSPV PNKDYEYIPS SLQPQYSAQL RRVLDEEPAP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
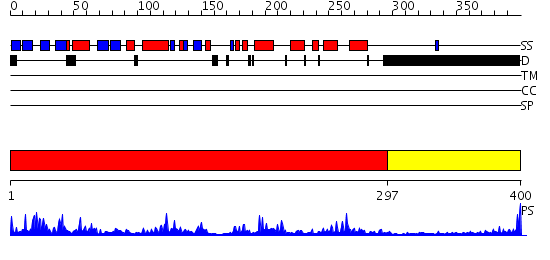
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..296] | 67.30103 | Casein kinase-1, CK1 | |
| 2 | View Details | [297..400] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)