













| General Information: |
|
| Name(s) found: |
csn1 /
SPBC215.03c
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 422 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA signalosome [TAS |
| Biological Process: |
regulation of meiotic cell cycle
[IMP protein catabolic process [IGI regulation of meiosis [IMP regulation of protein deneddylation [IMP regulation of DNA replication [IMP response to DNA damage stimulus [IMP regulation of ubiquitin-protein ligase activity [NAS regulation of proteasomal ubiquitin-dependent protein catabolic process [NAS] cullin deneddylation [NAS] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLNLLVNQE ELKRYLDEYG VWSKIFRALF VARNSKPLRS FCVHYAIKEL KEKTYNLELY 60
61 QSLFEEFQDC FENEQLDVEW VESVTFHRKQ NLEQLRRELK AYKNNLIRES IRAAQLDLAS 120
121 FFADVGQFDS ALRSYAKVRE YCTNAGQIAH LSLELMRISI WIGNYSHVLA FGSRAKSTVS 180
181 AAMELTSPIY AYCGLANFCL GDYEEALAHF LKVETDTSDG IITKTDISVY ISLCALACWD 240
241 HKRLIQELER NEEFNALSDL DPSLRRCLQA KCNRKYSLLL DTLQQNAQDY SLDMYLAPQL 300
301 TNLFSLIRER SLLDYLIPYS ALPFSKIAVD FHIDENFIEK NLLEIIEAKK LNGKVDSLQK 360
361 RVYIEPSSEP ENFEDIKNIA QTSLLYSKAL YLQTLAAMNT ENTDDEAPVI AQTNITAEDS 420
421 QN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
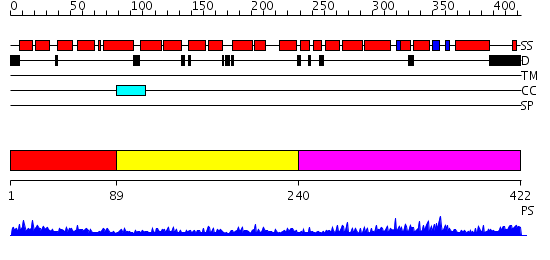
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | 6.315996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [89..239] | 6.522879 | Designed protein cTPR3 | |
| 3 | View Details | [240..422] | 11.69897 | Solution structure of the PCI domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)