













| General Information: |
|
| Name(s) found: |
FBpp0309258
[FlyBase]
nod-PA / FBpp0073363 [FlyBase] |
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 666 amino acids |
Gene Ontology: |
|
| Cellular Component: |
kinesin complex
[ISS]
|
| Biological Process: |
distributive segregation
[IMP]
DNA repair [IEA] mitotic spindle organization [IMP] meiotic chromosome segregation [IMP] establishment of meiotic spindle orientation [IMP] spindle assembly involved in female meiosis I [IMP] microtubule-based movement [IEA][ISS] positive regulation of microtubule polymerization [IDA] |
| Molecular Function: |
DNA binding
[IEA]
ATP binding [IEA] microtubule motor activity [IMP][ISS] microtubule plus-end binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEGAKLSAVR IAVREAPYRQ FLGRREPSVV QFPPWSDGKS LIVDQNEFHF DHAFPATISQ 60
61 DEMYQALILP LVDKLLEGFQ CTALAYGQTG TGKSYSMGMT PPGEILPEHL GILPRALGDI 120
121 FERVTARQEN NKDAIQVYAS FIEIYNEKPF DLLGSTPHMP MVAARCQRCT CLPLHSQADL 180
181 HHILELGTRN RRVRPTNMNS NSSRSHAIVT IHVKSKTHHS RMNIVDLAGS EGVRRTGHEG 240
241 VARQEGVNIN LGLLSINKVV MSMAAGHTVI PYRDSVLTTV LQASLTAQSY LTFLACISPH 300
301 QCDLSETLST LRFGTSAKKL RLNPMQVARQ KQSLAARTTH VFRQALCTST AIKSNAANHN 360
361 SIVVPKSKYS TTKPLSAVLH RTRSELGMTP KAKKRARELL ELEETTLELS SIHIQDSSLS 420
421 LLGFHSDSDK DRHLMPPPTG QEPRQASSQN STLMGIVEET EPKESSKVQQ SMVAPTVPTT 480
481 VRCQLFNTTI SPISLRASSS QRELSGIQPM EETVVASPQQ PCLRRSVRLA SSMRSQNYGA 540
541 IPKVMNLRRS TRLAGIREHA TSVVVKNETD AIPHLRSTVQ KKRTRNVKPA PKAWMANNTK 600
601 CFLDLLNNGN VKQLQEIPGI GPKSAFSLAL HRSRLGCFEN LFQVKSLPIW SGNKWERFCQ 660
661 INCLDT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
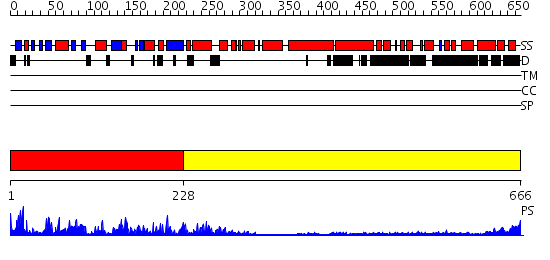
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..227] | 87.0 | Kinesin | |
| 2 | View Details | [228..666] | 13.69897 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)