













| General Information: |
|
| Name(s) found: |
FBpp0306615
[FlyBase]
dco-PC / FBpp0085106 [FlyBase] dco-PB / FBpp0085105 [FlyBase] dco-PA / FBpp0085104 [FlyBase] |
| Description(s) found:
Found 58 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 440 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][TAS]
cytoplasm [IDA][TAS] plasma membrane [IDA] |
| Biological Process: |
eclosion rhythm
[TAS]
regulation of protein import into nucleus [TAS] cell communication [IMP] establishment of ommatidial planar polarity [IMP] circadian rhythm [IMP][IPI][NAS][TAS] protein amino acid phosphorylation [IDA][NAS][TAS] regulation of ecdysteroid secretion [IMP] imaginal disc growth [IMP] rhythmic behavior [TAS] positive regulation of cell growth [IMP] behavioral response to cocaine [IMP][NAS] positive regulation of cell division [IMP] locomotor rhythm [NAS] negative regulation of apoptosis [IMP] establishment of imaginal disc-derived wing hair orientation [IMP] Wnt receptor signaling pathway [IMP] sleep [NAS] |
| Molecular Function: |
ATP binding
[IEA]
protein serine/threonine kinase activity [IEA][ISS][NAS] kinase activity [IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MELRVGNKYR LGRKIGSGSF GDIYLGTTIN TGEEVAIKLE CIRTKHPQLH IESKFYKTMQ 60
61 GGIGIPRIIW CGSEGDYNVM VMELLGPSLE DLFNFCSRRF SLKTVLLLAD QMISRIDYIH 120
121 SRDFIHRDIK PDNFLMGLGK KGNLVYIIDF GLAKKFRDAR SLKHIPYREN KNLTGTARYA 180
181 SINTHLGIEQ SRRDDLESLG YVLMYFNLGA LPWQGLKAAN KRQKYERISE KKLSTSIVVL 240
241 CKGFPSEFVN YLNFCRQMHF DQRPDYCHLR KLFRNLFHRL GFTYDYVFDW NLLKFGGPRN 300
301 PQAIQQAQDG ADGQAGHDAV AAAAAVAAAA AASSHQQQQH KVNAALGGGG GSAAQQQLQG 360
361 GQTLAMLGGN GGGNGSQLIG GNGLNMDDSM AATNSSRPPY DTPERRPSIR MRQGGGGGGG 420
421 GVGVGGMPSG GGGGGVGNAK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
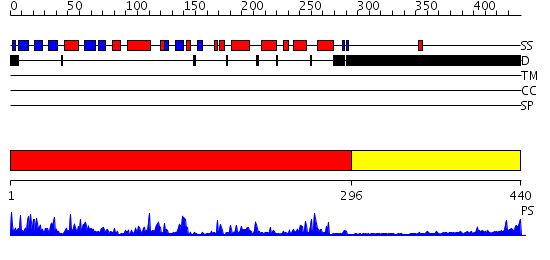
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..295] | 70.69897 | Casein kinase-1, CK1 | |
| 2 | View Details | [296..440] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.51 |
Source: Reynolds et al. (2008)