













| General Information: |
|
| Name(s) found: |
sqd-PB /
FBpp0082320
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 344 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC][IDA][TAS]
polytene chromosome puff [IDA] ribonucleoprotein complex [ISS][NAS] omega speckle [IDA] catalytic step 2 spliceosome [IDA] cytoplasm [TAS] precatalytic spliceosome [IDA] [NOT]nucleolus [IDA] chromatin [IDA] |
| Biological Process: |
germarium-derived egg chamber formation
[IMP]
intracellular mRNA localization [NAS][TAS] ovarian follicle cell migration [IMP] dorsal/ventral axis specification, ovarian follicular epithelium [IMP] dorsal/ventral pattern formation [IMP][TAS] oocyte anterior/posterior axis specification [IMP] nuclear mRNA splicing, via spliceosome [IC] mRNA export from nucleus [NAS] pole plasm mRNA localization [IMP] RNA export from nucleus [TAS] nuclear-transcribed mRNA catabolic process, nonsense-mediated decay [TAS] oogenesis [IMP][TAS] regulation of alternative nuclear mRNA splicing, via spliceosome [IMP] pole plasm oskar mRNA localization [IEP] negative regulation of RNA splicing [IMP] negative regulation of translation [TAS] oocyte microtubule cytoskeleton organization [IMP] oocyte localization during germarium-derived egg chamber formation [IMP] |
| Molecular Function: |
nucleic acid binding
[IEA]
nucleotide binding [IEA] SUMO binding [IPI] mRNA binding [ISS][NAS] mRNA 3'-UTR binding [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAENKQVDTE INGEDFTKDV TADGPGSENG DAGAAGSTNG SSDNQSAASG QRDDDRKLFV 60
61 GGLSWETTEK ELRDHFGKYG EIESINVKTD PQTGRSRGFA FIVFTNTEAI DKVSAADEHI 120
121 INSKKVDPKK AKARHGKIFV GGLTTEISDE EIKTYFGQFG NIVEVEMPFD KQKSQRKGFC 180
181 FITFDSEQVV TDLLKTPKQK IAGKEVDVKR ATPKPENQMM GGMRGGPRGG MRGGRGGYGG 240
241 RGGYNNQWDG QGSYGGYGGG YGGYGAGGYG DYYAGGYYNG YDYGYDGYGY GGGFEGNGYG 300
301 GGGGGNMGGG RGGPRGGGGP KGGGGFNGGK QRGGGGRQQR HQPY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
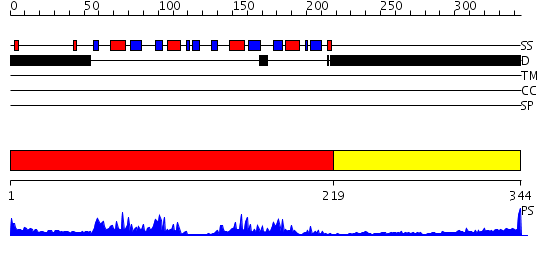
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..218] | 50.522879 | HNRNP A1 (RBD1,2) FROM HOMO SAPIENS | |
| 2 | View Details | [219..344] | 1.7 | Crystal structure of the C-terminal domain of BclA, the major antigen of the exosporium of the Bacillus anthracis spore. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)