













| General Information: |
|
| Name(s) found: |
Rel-PB /
FBpp0088973
[FlyBase]
Rel-PC / FBpp0088971 [FlyBase] Rel-PA / FBpp0088375 [FlyBase] |
| Description(s) found:
Found 52 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 971 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] |
| Biological Process: |
Toll signaling pathway
[TAS]
positive regulation of nitric oxide biosynthetic process [IMP] defense response to Gram-negative bacterium [IMP][TAS] signal transduction [TAS] cellular response to amino acid starvation [IMP] regulation of transcription, DNA-dependent [IEA] positive regulation of antibacterial peptide biosynthetic process [IEP][IMP][NAS][TAS] regulation of transcription [IEA] positive regulation of transcription from RNA polymerase II promoter [TAS] positive regulation of biosynthetic process of antibacterial peptides active against Gram-negative bacteria [IMP] positive regulation of antifungal peptide biosynthetic process [IMP] response to bacterium [TAS] immune response [IEP][IMP][TAS] positive regulation of innate immune response [IMP] innate immune response [IDA][NAS] positive regulation of gene expression [IMP] antimicrobial humoral response [IMP] positive regulation of gene-specific transcription from RNA polymerase II promoter [IDA] |
| Molecular Function: |
DNA binding
[IDA]
transcription factor activity [IEA][NAS][TAS] protein binding [IPI] sequence-specific DNA binding [IDA] transcription activator activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNMNQYYDLD NGKNVMFMND ASSTSGYSSS TSPNSTNRSF SPAHSPKTME LQTDFANLNL 60
61 PGGNSPHQPP MANSPYQNQL LNNGGICQLG ATNLINSTGV SFGVANVTSF GNMYMDHQYF 120
121 VPAPATVPPS QNFGYHQNGL ASDGDIKHVP QLRIVEQPVE KFRFRYKSEM HGTHGSLNGA 180
181 NSKRTPKTFP EVTLCNYDGP AVIRCSLFQT NLDSPHSHQL VVRKDDRDVC DPHDLHVSKE 240
241 RGYVAQFINM GIIHTAKKYI FEELCKKKQD RLVFQMNRRE LSHKQLQELH QETEREAKDM 300
301 NLNQVRLCFE AFKIEDNGAW VPLAPPVYSN AINNRKSAQT GELRIVRLSK PTGGVMGNDE 360
361 LILLVEKVSK KNIKVRFFEE DEDGETVWEA YAKFRESDVH HQYAIVCQTP PYKDKDVDRE 420
421 VNVYIELIRP SDDERSFPAL PFRYKPRSVI VSRKRRRTGS SANSSSSGTE SSNNSLDLPK 480
481 TLGLAQPPNG LPNLSQHDQT ISEEFGREKH LNEFIASEDF RKLIEHNSSD LEKICQLDMG 540
541 ELQHDGHNRA EVPSHRNRTI KCLDDLFEIY KQDRISPIKI SHHKVEKWFI EHALNNYNRD 600
601 TLLHEVISHK KDKLKLAIQT IQVMNYFNLK DVVNSTLNAD GDSALHVACQ QDRAHYIRPL 660
661 LGMGCNPNLK NNAGNTPLHV AVKEEHLSCV ESFLNGVPTV QLDLSLTNDD GLTPLHMAIR 720
721 QNKYDVAKKL ISYDRTSISV ANTMDGNNAL HMAVLEQSVE LLVLILDAQN ENLTDILQAQ 780
781 NAAGHTPLEL AERKANDRVV QLLKNVYPEK GELAMTWIPC KVKEEIDSSS DESSDAGQLE 840
841 IKSEEMDIET KDEDSVELDL SSGPRRQKDE SSRDTEMDNN KLQLLLKNKF IYDRLCSLLN 900
901 QPLGHGSDPQ DRKWMQLARQ THLKQFAFIW LGAEDLLDHV KRKGASVEFS TFARALQAVD 960
961 PQAYALLVNP T |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
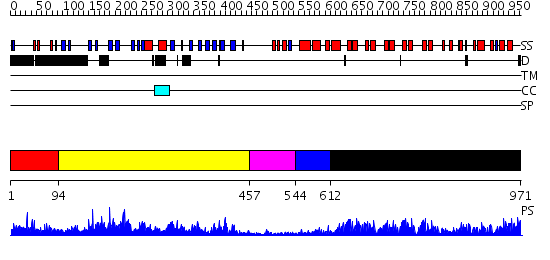
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [94..456] | 108.0 | p50 subunit of NF-kappa B transcription factor; p50 subunit of NF-kappa B (NFKB), N-terminal domain | |
| 3 | View Details | [457..543] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [544..611] | 9.69897 | Swi6 ankyrin-repeat fragment | |
| 5 | View Details | [612..971] | 83.045757 | Ankyrin-R |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)