













| General Information: |
|
| Name(s) found: |
Khc-PA /
FBpp0086328
[FlyBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 975 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA]
kinesin complex [IDA][ISS][NAS] |
| Biological Process: |
axon cargo transport
[IMP]
oocyte microtubule cytoskeleton polarization [IMP] oocyte dorsal/ventral axis specification [IMP] microtubule-based movement [IDA][ISS][NAS] pole plasm assembly [IMP] neurotransmitter secretion [NAS] cytoplasmic transport, nurse cell to oocyte [TAS] regulation of pole plasm oskar mRNA localization [IMP][TAS] eye photoreceptor cell differentiation [IGI] mitochondrion transport along microtubule [IMP] mitochondrion distribution [IMP] pole plasm oskar mRNA localization [NAS] cytoplasmic transport [IMP] neuronal action potential propagation [NAS] |
| Molecular Function: |
ATP binding
[IEA]
microtubule motor activity [IDA][ISS][NAS] plus-end-directed microtubule motor activity [TAS] motor activity [NAS] microtubule binding [IDA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAEREIPAE DSIKVVCRFR PLNDSEEKAG SKFVVKFPNN VEENCISIAG KVYLFDKVFK 60
61 PNASQEKVYN EAAKSIVTDV LAGYNGTIFA YGQTSSGKTH TMEGVIGDSV KQGIIPRIVN 120
121 DIFNHIYAME VNLEFHIKVS YYEIYMDKIR DLLDVSKVNL SVHEDKNRVP YVKGATERFV 180
181 SSPEDVFEVI EEGKSNRHIA VTNMNEHSSR SHSVFLINVK QENLENQKKL SGKLYLVDLA 240
241 GSEKVSKTGA EGTVLDEAKN INKSLSALGN VISALADGNK THIPYRDSKL TRILQESLGG 300
301 NARTTIVICC SPASFNESET KSTLDFGRRA KTVKNVVCVN EELTAEEWKR RYEKEKEKNA 360
361 RLKGKVEKLE IELARWRAGE TVKAEEQINM EDLMEASTPN LEVEAAQTAA AEAALAAQRT 420
421 ALANMSASVA VNEQARLATE CERLYQQLDD KDEEINQQSQ YAEQLKEQVM EQEELIANAR 480
481 REYETLQSEM ARIQQENESA KEEVKEVLQA LEELAVNYDQ KSQEIDNKNK DIDALNEELQ 540
541 QKQSVFNAAS TELQQLKDMS SHQKKRITEM LTNLLRDLGE VGQAIAPGES SIDLKMSALA 600
601 GTDASKVEED FTMARLFISK MKTEAKNIAQ RCSNMETQQA DSNKKISEYE KDLGEYRLLI 660
661 SQHEARMKSL QESMREAENK KRTLEEQIDS LREECAKLKA AEHVSAVNAE EKQRAEELRS 720
721 MFDSQMDELR EAHTRQVSEL RDEIAAKQHE MDEMKDVHQK LLLAHQQMTA DYEKVRQEDA 780
781 EKSSELQNII LTNERREQAR KDLKGLEDTV AKELQTLHNL RKLFVQDLQQ RIRKNVVNEE 840
841 SEEDGGSLAQ KQKISFLENN LDQLTKVHKQ LVRDNADLRC ELPKLEKRLR CTMERVKALE 900
901 TALKEAKEGA MRDRKRYQYE VDRIKEAVRQ KHLGRRGPQA QIAKPIRSGQ GAIAIRGGGA 960
961 VGGPSPLAQV NPVNS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
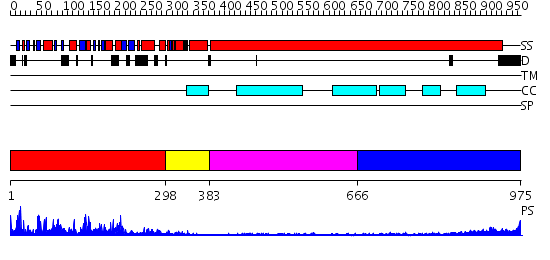
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..297] | 87.522879 | Kinesin | |
| 2 | View Details | [298..382] | 10.0 | KINESIN (DIMERIC) FROM RATTUS NORVEGICUS | |
| 3 | View Details | [383..665] | 2.84 | Tropomyosin | |
| 4 | View Details | [666..975] | 5.30103 | No description for 2i1jA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)