













| General Information: |
|
| Name(s) found: |
ASR1 /
YPR093C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 288 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
response to ethanol
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEECPICLAD DQEGEQFGCL NVCGHKFHLN CIREWHKYSI NLKCPICRVE STHLEVGEGQ 60
61 HALSINLKMG FMIKNAIDYV GAETTNERNE DDTGEQDQEI EFLSERLRGT LVMDTIKIIQ 120
121 CSICGDTDVS RLSLYCQDCE AIYHETCLRG LACEVGDRNT WQECTDCRSN ALLELRMGAI 180
181 SSQLASYDSR NSMIFAGELR DKHSVKTQQM YEQIRNAKHK IQMHVRRALD RYPLPLLRFK 240
241 DAYKHVNKQV SRKLYRLSDN KYLPDQYDYD SLARTGVHTE LLIYCHDE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ASR1 RPB2 RPO21 |
| View Details | Krogan NJ, et al. (2006) |

|
ARC15 ASR1 CAJ1 CDC7 CRN1 INP52 IRC22 MSK1 PNO1 PST2 RAD34 SAC6 SAR1 SLM1 TCP1 TED1 YPP1 |
| View Details | Ho Y, et al. (2002) |
|
ASR1 FPR1 RPB11 RPB3 RPB9 RPO21 YDR131C |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
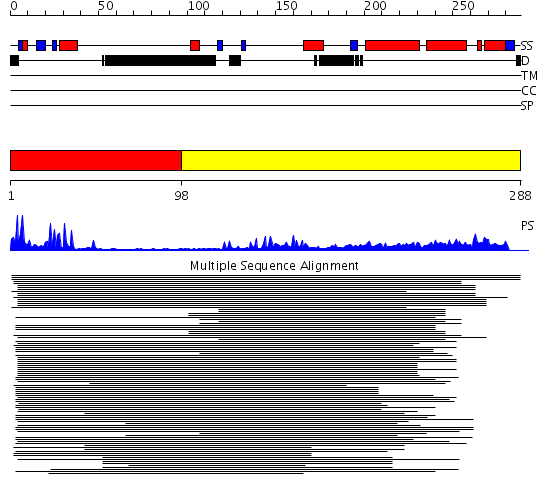
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..97] | 202.392997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [98..288] | 9.274996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [178..211] | 3.490996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [212..288] | 1.275992 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)