













| General Information: |
|
| Name(s) found: |
SWH1 /
YAR042W
[SGD]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1188 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear envelope
[IDA early endosome [IDA endoplasmic reticulum [IDA Golgi trans cisterna [IDA |
| Biological Process: |
sterol transport
[IGI endocytosis [IGI exocytosis [IGI |
| Molecular Function: |
oxysterol binding
[ISS phosphatidylinositol binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEQPDLSSVA ISKPLLKLKL LDALRQGSFP NLQDLLKKQF QPLDDPNVQQ VLHLMLHYAV 60
61 QVAPMAVIKE IVHHWVSTTN TTFLNIHLDL NERDSNGNTP LHIAAYQSRG DIVAFLLDQP 120
121 TINDCVLNNS HLQAIEMCKN LNIAQMMQVK RSTYVAETAQ EFRTAFNNRD FGHLESILSS 180
181 PRNAELLDIN GMDPETGDTV LHEFVKKRDV IMCRWLLEHG ADPFKRDRKG KLPIELVRKV 240
241 NENDTATSTK IAIDIELKKL LERATREQSV IDVTNNNLHE APTYKGYLKK WTNFAQGYKL 300
301 RWFILSSDGK LSYYIDQADT KNACRGSLNM SSCSLHLDSS EKLKFEIIGG NNGVIRWHLK 360
361 GNHPIETNRW VWAIQGAIRY AKDREILLHN GPYSPSLALS HGLSSKVSNK ENLHATSKRL 420
421 TKSPHLSKST LTQNDHDNDD DSTNNNNNKS NNDYDDNNNN NNNDDDDYDD DDESRPLIEP 480
481 LPLISSRSQS LSEITPGPHS RKSTVSSTRA ADIPSDDEGY SEDDSDDDGN SSYTMENGGE 540
541 NDGDEDLNAI YGPYIQKLHM LQRSISIELA SLNELLQDKQ QHDEYWNTVN TSIETVSEFF 600
601 DKLNRLTSQR EKRMIAQMTK QRDVNNVWIQ SVKDLEMELV DKDEKLVALD KERKNLKKML 660
661 QKKLNNQPQV ETEANEESDD ANSMIKGSQE STNTLEEIVK FIEATKESDE DSDADEFFDA 720
721 EEAASDKKAN DSEDLTTNKE TPANAKPQEE APEDESLIVI SSPQVEKKNQ LLKEGSFVGY 780
781 EDPVRTKLAL DEDNRPKIGL WSVLKSMVGQ DLTKLTLPVS FNEPTSLLQR VSEDIEYSHI 840
841 LDQAATFEDS SLRMLYVAAF TASMYASTTN RVSKPFNPLL GETFEYARTD GQYRFFTEQV 900
901 SHHPPISATW TESPKWDFYG ECNVDSSFNG RTFAVQHLGL WYITIRPDHN ISVPEETYSW 960
961 KKPNNTVIGI LMGKPQVDNS GDVKVTNHTT GDYCMLHYKA HGWTSAGAYE VRGEVFNKDD1020
1021 KKLWVLGGHW NDSIYGKKVT ARGGELTLDR IKTANSATGG PKLDGSKFLI WKANERPSVP1080
1081 FNLTSFALTL NALPPHLIPY LAPTDSRLRP DQRAMENGEY DKAAAEKHRV EVKQRAAKKE1140
1141 REQKGEEYRP KWFVQEEHPV TKSLYWKFNG EYWNKRKNHD FKDCADIF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
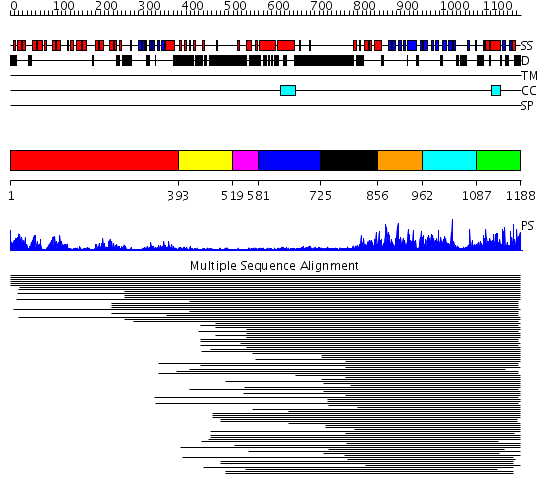
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..392] | 60.0 | Ankyrin-R | |
| 2 | View Details | [393..518] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [519..580] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [581..724] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [725..855] | 1.180993 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [856..961] | 1.143975 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [962..1086] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1087..1188] | 3.462992 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)