













| General Information: |
|
| Name(s) found: |
PHR1 /
YOR386W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 565 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS mitochondrion [IDA |
| Biological Process: |
photoreactive repair
[TAS |
| Molecular Function: |
deoxyribodipyrimidine photo-lyase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRTVISSSN AYASKRSRLD IEHDFEQYHS LNKKYYPRPI TRTGANQFNN KSRAKPMEIV 60
61 EKLQKKQKTS FENVSTVMHW FRNDLRLYDN VGLYKSVALF QQLRQKNAKA KLYAVYVINE 120
121 DDWRAHMDSG WKLMFIMGAL KNLQQSLAEL HIPLLLWEFH TPKSTLSNSK EFVEFFKEKC 180
181 MNVSSGTGTI ITANIEYQTD ELYRDIRLLE NEDHRLQLKY YHDSCIVAPG LITTDRGTNY 240
241 SVFTPWYKKW VLYVNNYKKS TSEICHLHII EPLKYNETFE LKPFQYSLPD EFLQYIPKSK 300
301 WCLPDVSEEA ALSRLKDFLG TKSSKYNNEK DMLYLGGTSG LSVYITTGRI STRLIVNQAF 360
361 QSCNGQIMSK ALKDNSSTQN FIKEVAWRDF YRHCMCNWPY TSMGMPYRLD TLDIKWENNP 420
421 VAFEKWCTGN TGIPIVDAIM RKLLYTGYIN NRSRMITASF LSKNLLIDWR WGERWFMKHL 480
481 IDGDSSSNVG GWGFCSSTGI DAQPYFRVFN MDIQAKKYDP QMIFVKQWVP ELISSENKRP 540
541 ENYPKPLVDL KHSRERALKV YKDAM |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
CDC55 FIR1 PHR1 PPH21 PPH22 RRD2 TPD3 YRR1 ZDS1 ZDS2 |
| View Details | Ho Y, et al. (2002) |

|
MSD1 PHR1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
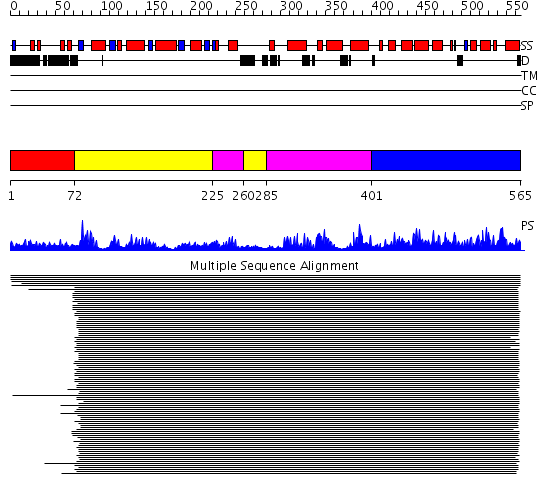
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..71] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [72..224] [260..284] |
809.0103 | C-terminal domain of DNA photolyase; DNA photolyase | |
| 3 | View Details | [225..259] [285..400] |
809.0103 | C-terminal domain of DNA photolyase; DNA photolyase | |
| 4 | View Details | [401..565] | 809.0103 | C-terminal domain of DNA photolyase; DNA photolyase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)