













| General Information: |
|
| Name(s) found: |
PKH2 /
YOL100W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1081 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
MAPKKK cascade involved in cell wall biogenesis
[IGI protein amino acid phosphorylation [IDA |
| Molecular Function: |
protein kinase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYFDKDNSMS PRPLLPSDEQ KLNINLLTKK EKFSHLDPHY DAKATPQRST SNRNVGDLLL 60
61 EKRTAKPMIQ KALTNTDNFI EMYHNQQRKN LDDDTIKEVM INDENGKTVA STNDGRYDND 120
121 YDNNDINDQK TLDNIAGSPH MEKNRNKVKI EHDSSSQKPI AKESSKAQKN IIKKGIKDFK 180
181 FGSVIGDGAY STVMLATSID TKKRYAAKVL NKEYLIRQKK VKYVSIEKTA LQKLNNSPSV 240
241 VRLFSTFQDE SSLYFLLEYA PNGDFLSLMK KYGSLDETCA RYYAAQIIDA IDYLHSNGII 300
301 HRDIKPENIL LDGEMKIKLT DFGTAKLLNP TNNSVSKPEY DLSTRSKSFV GTAEYVSPEL 360
361 LNDSFTDYRC DIWAFGCILF QMIAGKPPFK ATNEYLTFQK VMKVQYAFTP GFPLIIRDLV 420
421 KKILVKNLDR RLTISQIKEH HFFKDLNFKD GSVWSKTPPE IKPYKINAKS MQAMPSGSDR 480
481 KLVKKSVNTL GKSHLVTQRS ASSPSVEETT HSTLYNNNTH ASTESEISIK KRPTDERTAQ 540
541 ILENARKGIN NRKNQPGKRT PSGAASAALA ASAALTKKTM QSYPTSSSKS SRSSSPATTS 600
601 RPGTYKRTSS TESKPFAKSP PLSASVLSSK VPMPPYTPPM SPPMTPYDTY QMTPPYTTKQ 660
661 QDYSDTAIAA PKPCISKQNV KNSTDSPLMN KQDIQWSFYL KNINEHVLRT EKLDFVTTNY 720
721 DILEKKMLKL NGSLLDPQLF GKPRHTFLSQ VARSGGEVTG FRNDPTMTAY SKTEDTYYSK 780
781 NIIDLQLLED DYRIEGGDLS ELLTNRSGEG YKCNQNSSPM KDDDKSESNN KGSSVFSGKI 840
841 KKLFHPTSAA ETLSSSDEKT KYYKRTIVMT SFGRFLVFAK RRQPNPVTNL KYELEYDINL 900
901 RQQGTKIKEL IIPLEMGTNH IVVIQTPYKS FLLSTDKKTT SKLFTVLKKI LNSNTNKIEK 960
961 ELLQRNQKVI ERRTSSSGRA IPKDLPTSKS PSPKPRTHSQ SPSISKHNSF SESINSAKSN1020
1021 RSSRIFETFI NAKEQNSKKH AAPVPLTSKL VNGLPKRQVT VGLGLNTGTN FKNSSAKSKR1080
1081 S |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
|
HXT6 HXT7 LSP1 PIL1 PKH2 TIM21 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
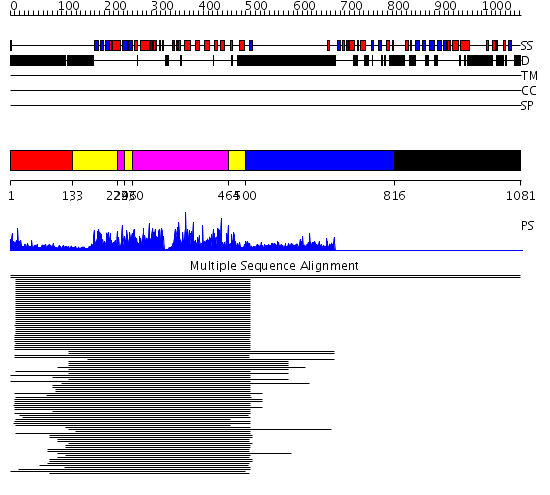
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..132] | 15.522879 | C2 domain from protein kinase c (beta) | |
| 2 | View Details | [133..228] [243..259] [464..499] |
573.9897 | cAMP-dependent PK, catalytic subunit | |
| 3 | View Details | [229..242] [260..463] |
573.9897 | cAMP-dependent PK, catalytic subunit | |
| 4 | View Details | [500..815] | 13.24 | RBP1 | |
| 5 | View Details | [816..1081] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)