













| General Information: |
|
| Name(s) found: |
RKR1 /
YMR247C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1562 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA ribosome [IDA |
| Biological Process: |
chromatin modification
[IGI chromatin silencing at telomere [IMP protein ubiquitination [IDA |
| Molecular Function: |
ubiquitin-protein ligase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFGGINTFQ QYNTDLGLGH NGVRISLNYF DGLPDPSLLN SLYSNELKLI FKSLLKRDET 60
61 TKEKALMDLS NLISDFNQNE YFFNDIFLLC WSQIYAKLII SDYKVIRLQS HQITIMLVKS 120
121 LRKKISKFLK DFIPLILLGT CELDYSVSKP SLNELTECFN KDPAKINALW AVFQEQLLNL 180
181 VKEIVVNENE DTISDERYSS KEESEFRYHR VIASAVLLLI KLFVHNKDVS ERNSSSLKVI 240
241 LSDESIWKLL NLKNGQNTNA YETVLRLIDV LYTRGYMPSH KNIMKLAVKK LLKSLTHITS 300
301 KNILKVCPVL PSILNLLATL DDYEDGTIWS YDKSSKEKVL KFLSVSRTSP SPGFFNAVFA 360
361 LYSSTKRHSF LDYYLEWLPF WQKSVQRLNE KGFSARNSAE VLNEFWTNFL KFAEDSSEER 420
421 VKKMVESEIF NSLSCGKSLS EYTKLNQTLS GVFPPDKWER EIEDYFTSDE DIRKIKVSFE 480
481 KNLFALLVTS PNNESAISRL FDFFVQLIET DPSNVFNKYD GVYDALNYFL DSDMIFLNGK 540
541 IGKFINEIPT LVQESTYQNF AGIMAQYSNS KFFKMNTDAI TSLEDFFIVA LSFNLPKTII 600
601 LATMNELDND IYQQLMKSDS LELELYIEDF MKNYKFDDSG EIFKGNNKFL NQRTITTLYR 660
661 SAVANGQVEQ FCAVLSKLDE TFFSTLLLNT DFLSCALYEV SEDTNEKLFK LSLQLAKGNS 720
721 EIANKLAQVI LQHAQVYFSP GAKEKYVTHA VELINGCNDT SQIFFPANAI EVFARYMPAI 780
781 DYRSSLVSSL STNTHLLLTD DKPINLKNMQ KLIRYALFLD ALLDALPERV NNHIVAFITV 840
841 VSELVTDYNC LSEEPNDLYY DFGHTFFKHG KVNLNFSDIV GNVIQPANGG DAMLTFDIAE 900
901 SNSVYFFYYS RVLYKVLLNS IDTVSSTTLN GLLASVESFV TKTVRDQKST DKDYLLCAIL 960
961 LLMFNRSNSK DEITKLRTLL ASQLIGIREV ELVDQEFKSL ALLNNLLDIP QADKQFVPIA1020
1021 PQRLNMIFRS ILKWLDSDLA YEPSFSTVRL LLLDFFTKLM RFEGVRDMGI TAFELSERLL1080
1081 ADSLSMCQID DTLYLLELRS SCLNLYETLS QGVSKNGEEI SEYGDEIQEN LIELMFLNFN1140
1141 QERNNQVSTL FYQKLYKVIS SMELKKLESQ YKRIFEVVLN DKDIGSNINQ SRLLTTLLGS1200
1201 LVVKTQQDII IEYELRIQKQ TGSDVDGSAS DNDVNSKFKL PQKLLQKVTD EVPKEYLEYE1260
1261 NKNSFIKYLW YWHLILMYFK DTSYNMRQIF IEQLKEAGLI NRMFDFITDQ IDLRDTEFWK1320
1321 QVDTKEISEY NIVGNNFSPY KEDIFEECKK LLGHTLYQLF NNVGCLTSIW WLNIKDRTLQ1380
1381 NDIEKFVSEF ISPILIKNEF DDINSKMDRL TSNDDALTIK LNNITNEVKA SYLIDDQKLE1440
1441 ISFKLPKNYP LTNIQVNGVS RVGISEQKWK QWIMSTQHVI TGMNGSVLDS LELFTKNVHL1500
1501 QFSGFEECAI CYSILHAVDR KLPSKTCPTC KNKFHGACLY KWFRSSGNNT CPLCRSEIPF1560
1561 RR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
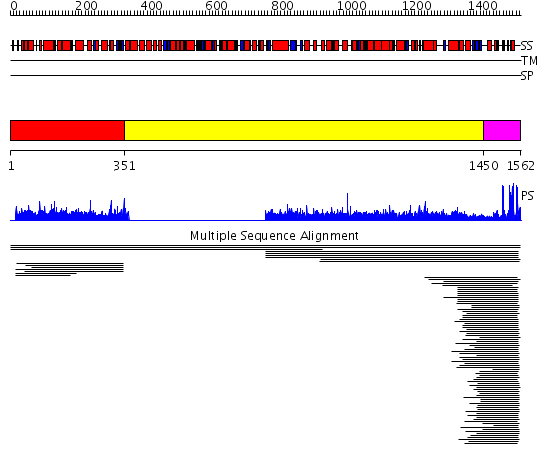
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..350] | 3.008922 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [351..1449] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [1450..1562] | 3.0 | Not-4 N-terminal RING finger domain | |
| 4 | View Details | [1015..1226] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1227..1480] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1481..1562] | 15.0 | Solution structure of RING finger in RING finger protein 38 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)