













| General Information: |
|
| Name(s) found: |
MNR2 /
YKL064W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 969 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[ISS |
| Biological Process: |
magnesium ion transport
[ISS |
| Molecular Function: |
magnesium ion transmembrane transporter activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTDNSQKDE GVPLLSPYSS SPQLRKKKRN QKRRKDKFVG HLKSDSRRPT QLLHDNLQHN 60
61 HGQITDFDQI DSWGMLHESD STSNDIIKSE DPSLKGAFID HRPSMSQPRE GPQSVSSTVQ 120
121 PQPIMKFSTP SYKKPAGLRP SDQNRSLVSD LSPSELESWL KRRKSVHKSF VDENSPTDRR 180
181 QSNANNDVVI DVDALMNHVN NNASTGVNDN SKRRKKKRGS DDSSNKNSKS TSSDSNDEED 240
241 EYNSRPSSSL SSNNSSLDDV CLVLDDEGSE VPKAWPDCTV LEEFSKEETE RLRSQAIQDA 300
301 EAFHFQYDED EEDGTSNEDG ILFSKPIVTN IDVPELGNRR VNETENLKNG RLRPKRIAPW 360
361 HLIQRPMVLG SNSTKDSKSR IQSGLQDNLL VGRNIQYPPH IISNNPEHFR FTYFRVDLDS 420
421 TVHSPTISGL LQPGQKFQDL FVASIYSQDN SAGHIKTHPN SPTPGIKAET VSQLQGLTAK 480
481 NPSTLSSMSV ANIEDVPPFW LDVSNPTEEE MKILSKAFGI HPLTTEDIFL GEVREKVELF 540
541 RDYYLICFRS FDIVAEKHVR RRRKEKQESA TLDHESISRR KSQAYGATMS NESNANNNNS 600
601 TSNASRSKWL PSILRARRRS SANRTTNTSS SSYKRRVKSE KKKMEENEKF KRKSGDRHKP 660
661 REGELEPLNV YIIVFRTGVL TFHFAPTPHP INVRRRARLL KDYLNVTSDW IAYALIDDIT 720
721 DAFAPMIELI EDEVYEIEDA ILKMHQSDDS SDSDSSDSDS DSGASDEDAF PFDVYSKKTS 780
781 YSSAKSSVSS RSMSTSEASF NANLIGWKRK GDMLRRIGEC RKRVMSILRL LGSKADVIKG 840
841 FAKRYNEQWE ASPQSEIAMY LGDIQDHIVT MVSSLNHYEK LLSRSHSNYL AQINIDMTKV 900
901 NNDMNDVLGK ITILGTIVLP MNVITGLWGM NVIVPGQYRD SLTWFIGIVL FMCMLACSAY 960
961 MYTKRRFGF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |

|
MNR2 MRS2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | hx21: both fusion and trans-SNARE complex formation take place. Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | hx23: mixture in detergent (control) Investigating Vam3p, Nyv1p and their partners in trans-SNARE complex | Hao Xu, et al. (2010) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
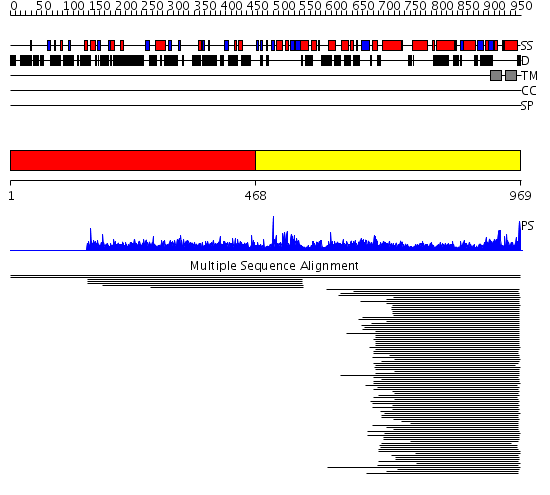
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..467] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [468..969] | 141.79588 | CorA-like Mg2+ transporter protein No confident structure predictions are available. | |
| 3 | View Details | [349..490] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [491..598] | 27.0 | No description for 2hn1A was found. | |
| 5 | View Details | [599..969] | 63.045757 | No description for 2iubA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 5 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|