













| General Information: |
|
| Name(s) found: |
RTT107 /
YHR154W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1070 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
negative regulation of transposition, DNA-mediated
[IMP double-strand break repair [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTSLLFEQL NFLILVAAEA ELPIAHSTRK LLMDNSCNNC QIYELYNENL KDVKTDKDWF 60
61 MNKFGPQTVH FVISNTINFP FYKIVYFDLL IPVVSHTWVQ DSVKTKRHLR TNMYSPNPFH 120
121 LLRDCQVYIS KSSFNKCEYI LYSDLLHLLG GTLVNYISNR TTHVIVQSPQ DPIIATVSKL 180
181 TFGSFSSSST NKHTEKPLRE WKFVYPIWIL YHFKMAKPLK GELATLCELD MQDTSEEQLF 240
241 AKWEEVIGDK QTSSSQLTLH PNKTLFKNHH FAISPDLNFF TPLYWFLKGF IEDLDGKVTP 300
301 LSFSDDLKSV YQAFPDIDCY IGHSANSPIL EKTKSIKPEI HVGNVSWLFY MFALQKFTPV 360
361 SQCKLIHQPF HAKLFTSKEL TVAYTNYFGS QRFYIQRLVE ILGGLSTPEL TRKNTHLITK 420
421 STIGKKFKVA KKWSLDPQNA IIVTNHMWLE QCYMNNSKLN PKDSRFQNFK LDDNMGWNIG 480
481 QIGMDHSSLP TPKNLSMVTY DTQSISEKPP PTNDQMDQIN DNTNVLSKKD GTPISSFENS 540
541 IDEKIDKLQK ISGEVAVTHS GDLERSFVSR PSRASFPVVD SKKSNLQKKD SNSDISMETE 600
601 VFCEGHEKRE EKEFTKPITE YDAPKKQEIR EQSRKKNDID YKKEEEETEL QVQLGQRTKR 660
661 EIKTSKKNEK EKETNECHIE VDQMTNEKQG EESTGKLIST EDVTSKKDTD KFSHLFEGLS 720
721 DNDDHINDEK PAVNSKYTTP KTSQNITSGV DTPTTAQTQV FMPSSGNSRL AKTQAAKRLH 780
781 TDIESLNEFQ KNFKRKRIDS EEISLSQDVE RSNNNKELAT KAEKILARFN ELPNYDLKAV 840
841 CTGCFHDGFN EVDIEILNQL GIKIFDNIKE TDKLNCIFAP KILRTEKFLK SLSFEPLKFA 900
901 LKPEFIIDLL KQIHSKKDKL SQININLFDY EINGINESII SKTKLPTKVF ERANIRCINL 960
961 VNDIPGGVDT IGSVLKAHGI EKINVLRSKK CTFEDIIPND VSKQENGGIF KYVLIVTKAS1020
1021 QVKKFTKLIN DRDKNETILI VEWNWCVESI FHLNVDFTSK KNVLYQKKNN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
DBP10 RTT107 |
| View Details | Ho Y, et al. (2002) |
|
GDH2 MMS22 RTT101 RTT107 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
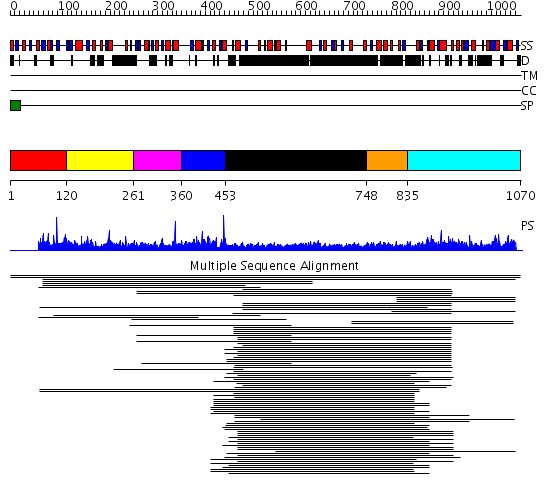
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..119] | 8.32 | Breast cancer associated protein, BRCA1 | |
| 2 | View Details | [120..260] | 8.32 | Breast cancer associated protein, BRCA1 | |
| 3 | View Details | [261..359] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [360..452] | 7.91 | NAD+-dependent DNA ligase, domain 3; NAD+-dependent DNA ligase; Adenylation domain of NAD+-dependent DNA ligase | |
| 5 | View Details | [453..747] | 32.39794 | Heavy meromyosin subfragment | |
| 6 | View Details | [748..834] | 4.30103 | Tropomyosin | |
| 7 | View Details | [835..1070] | 2.803997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|