













| General Information: |
|
| Name(s) found: |
ECM14 /
YHR132C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 430 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type vacuole
[IDA |
| Biological Process: |
proteolysis
[ISS cellular cell wall organization [IMP |
| Molecular Function: |
metalloendopeptidase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLHMNSLWGC FLFVLLAVTG AVQGLQEDYS EYAVYRFTSD NYSTLVRDVI APLTDDYDVW 60
61 TRSNNFIDIK LPKEIGEQIN DGQVIIDNMN ELIQNTLPTS QMMAREQAVF ENDYDFFFNE 120
121 YRDLDTIYMW LDLLERSFPS LVAVEHLGRT FEGRELKALH ISGNKPESNP EKKTIVITGG 180
181 IHAREWISVS TVCWALYQLL NRYGSSKKET KYLDDLDFLV IPVFNPDGYA YTWSHDRLWR 240
241 KNRQRTHVPQ CLGIDIDHSF GFQWEKAHTH ACSEEYSGET PFEAWEASAW YKYINETKGD 300
301 YKIYGYIDMH SYSQEILYPY AYSCDALPRD LENLLELSYG LSKAIRSKSG RNYDVISACK 360
361 DRGSDIFPGL GAGSALDFMY HHRAHWAFQL KLRDTGNHGF LLPPENIKPV GKETYAALKY 420
421 FCDFLLDPEI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |

|
ECM14 PRC1 VMA9 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
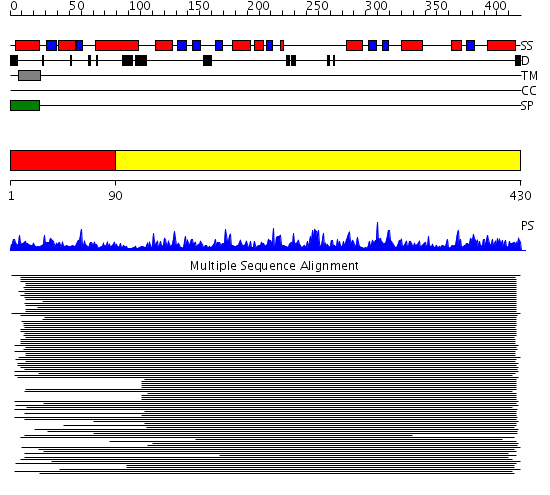
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | 583.0103 | Carboxypeptidase A; Procarboxypeptidase A | |
| 2 | View Details | [90..430] | 583.0103 | Carboxypeptidase A; Procarboxypeptidase A |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|