













| General Information: |
|
| Name(s) found: |
YHR035W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 630 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCSPTNFLYE PFSSDAVTQN YDQNLKCTKC GAYYSMACSL REQNVWTCLF CNQSNSNAEL 60
61 PLVPSNTYTL TSAKKEILSR RTIMIIDAIC DPHELNYLVS ILCNNYITRQ QEPLSIITIQ 120
121 QSGHVILHNA VNHRRDAVFS INEFMTKYNL DKLNASYFEK KISEINQESY WFDKSTQGSL 180
181 RKLLREICKI ANKVNISSKR DKRCTGLALF VSSVLASQCS LSAYCHIVSF LNGPCTKGGG 240
241 KVMSRERGES MRQNHHFESK SSQLQLSKSP TKFYKKMLEK FANQSLIYEF FIASLDQIGI 300
301 LEMSPLITSS MAVSQFDSFN DERFAMSFQK YLNLRDHNAI YNCHSKIMTA KNAIVVKDFP 360
361 KYSLNPKNLS LPLEISLGHN SAEAPIQFQT TFENQTEKYI RIETLLLPKA NRSFGAQNEI 420
421 VFSMKKIASR IIDSFAYSSK HTKELMKQLF LLPNQIRGKD VDMVNLIQWC YHIYRSPILS 480
481 VRNTSPDERY LFLHRIINAS KDTCLSLCKP FIWSYSDLKH DWIVLDVPLT RAQILQDDKT 540
541 TICVDGGSYL VLRRGKLLEK EGRELCCKLL NDLQRFPQPL YVETKTGGSQ DRFLKSKIIP 600
601 LDITDKETLG TEDMTFNEYF NLFTDLSGSK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
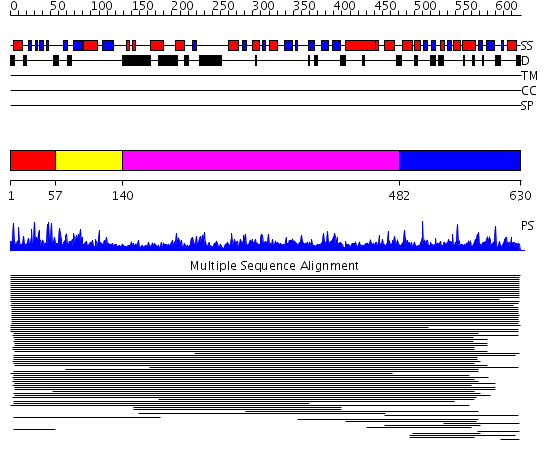
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..56] | 1.041985 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [57..139] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [140..481] | 1.049938 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [482..630] | 2.69897 | Severin, domain 2 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)