













| General Information: |
|
| Name(s) found: |
SLH1 /
YGR271W
[SGD]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1967 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
regulation of translation
[IGI |
| Molecular Function: |
RNA helicase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTEYSADSS KSFMIAMQSM IDTSQTFNLD RSKISLPDFD DELKKVQKDE PNQRTELTVL 60
61 SQDRNDWDDI FEEFKDISFA QLQSIIDSYK TKNAVAVYKK IGKLINEAET TLSSNVLLET 120
121 VLQMVYKHQK QELEKELLDF LGTGNIDLVS LLLQHRRMIV ATPIETTILL IKNAVNSTPE 180
181 FLTQQDIRNQ VLESAEDAKN RKLNPATKII KYPHVFRKYE AGSTTAMAFA GQKFTLPVGT 240
241 TRMSYNTHEE IIIPAADQAS NKNYLYTKLL KISDLDHFCK TVFPYETLNQ IQSLVYPVAY 300
301 KTNENMLICA PTGAGKTDIA LLTIINTIKQ FSVVNGENEI DIQYDDFKVI YVAPLKALAA 360
361 EIVDKFSKKL APFNIQVREL TGDMQLTKAE ILATQVIVTT PEKWDVVTRK ANGDNDLVSK 420
421 VKLLIIDEVH LLHEDRGPVI ETLVARTLRQ VESSQSMIRI IGLSATLPNF MDVADFLGVN 480
481 RQIGMFYFDQ SFRPKPLEQQ LLGCRGKAGS RQSKENIDKV AYDKLSEMIQ RGYQVMVFVH 540
541 SRKETVKSAR NFIKLAESNH EVDLFAPDPI EKDKYSRSLV KNRDKDMKEI FQFGFGIHHA 600
601 GMARSDRNLT EKMFKDGAIK VLCCTATLAW GVNLPADCVI IKGTQVYDSK KGGFIDLGIS 660
661 DVIQIFGRGG RPGFGSANGT GILCTSNDRL DHYVSLITQQ HPIESRFGSK LVDNLNAEIS 720
721 LGSVTNVDEA IEWLGYTYMF VRMRKNPFTY GIDWEEIAND PQLYERRRKM IVVAARRLHA 780
781 LQMIVFDEVS MHFIAKDLGR VSSDFYLLNE SVEIFNQMCD PRATEADVLS MISMSSEFDG 840
841 IKFREEESKE LKRLSDESVE CQIGSQLDTP QGKANVLLQA YISQTRIFDS ALSSDSNYVA 900
901 QNSVRICRAL FLIGVNRRWG KFSNVMLNIC KSIEKRLWAF DHPLCQFDLP ENIIRRIRDT 960
961 KPSMEHLLEL EADELGELVH NKKAGSRLYK ILSRFPKINI EAEIFPITTN VMRIHIALGP1020
1021 DFVWDSRIHG DAQFFWVFVE ESDKSQILHF EKFILNRRQL NNQHEMDFMI PLSDPLPPQV1080
1081 VVKVVSDTWI GCESTHAISF QHLIRPFNET LQTKLLKLRP LPTSALQNPL IESIYPFKYF1140
1141 NPMQTMTFYT LYNTNENAFV GSPTGSGKTI VAELAIWHAF KTFPGKKIVY IAPMKALVRE1200
1201 RVDDWRKKIT PVTGDKVVEL TGDSLPDPKD VHDATIVITT PEKFDGISRN WQTRKFVQDV1260
1261 SLIIMDEIHL LASDRGPILE MIVSRMNYIS SQTKQPVRLL GMSTAVSNAY DMAGWLGVKD1320
1321 HGLYNFPSSV RPVPLKMYID GFPDNLAFCP LMKTMNKPVF MAIKQHSPDK PALIFVASRR1380
1381 QTRLTALDLI HLCGMEDNPR RFLNIDDEEE LQYYLSQVTD DTLKLSLQFG IGLHHAGLVQ1440
1441 KDRSISHQLF QKNKIQILIA TSTLAWGVNL PAHLVIIKGT QFFDAKIEGY RDMDLTDILQ1500
1501 MMGRAGRPAY DTTGTAIVYT KESKKMFYKH FLNVGFPVES SLHKVLDDHL GAEITSGSIT1560
1561 NKQEALDFLS WTFLFRRAHH NPTYYGIEDD TSTAGVSEHL SSLIDSTLEN LRESQCVLLH1620
1621 GDDIVATPFL SISSYYYISH LTIRQLLKQI HDHATFQEVL RWLSLAVEYN ELPVRGGEII1680
1681 MNEEMSQQSR YSVESTFTDE FELPMWDPHV KTFLLLQAHL SRVDLPIADY IQDTVSVLDQ1740
1741 SLRILQAYID VASELGYFHT VLTMIKMMQC IKQGYWYEDD PVSVLPGLQL RRIKDYTFSE1800
1801 QGFIEMTPQQ KKKKLLTLEE IGRFGYKKLL NVFDQLTFGM TESEDTKKRF VSVCQRLPVL1860
1861 EGMKFEEQEN NEVLTFYSKH LSSKHNNGFE VYCDKFPKIQ KELWFLIGHK GDELLMIKRC1920
1921 QPKQMNKEVI IHCDLFIPEE IRGEELQFSL INDALGLRYD MVHKLIS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
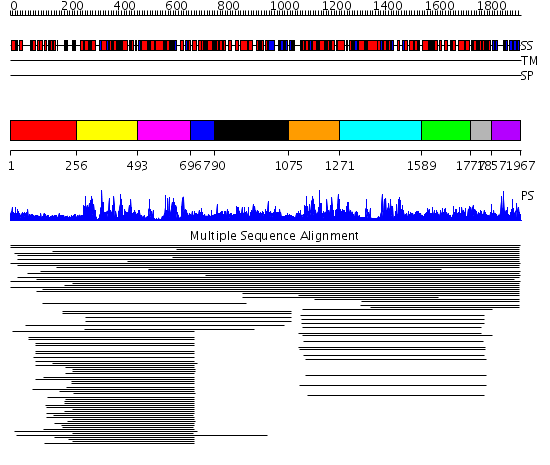
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..255] | 10.589997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [256..492] | 80.69897 | Putative DEAD box RNA helicase | |
| 3 | View Details | [493..695] | 80.69897 | Putative DEAD box RNA helicase | |
| 4 | View Details | [696..789] | 4.39794 | Nucleotide excision repair enzyme UvrB | |
| 5 | View Details | [790..1074] | 1.091995 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [1075..1270] | 14.93 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 7 | View Details | [1271..1588] | 6.0 | Nucleotide excision repair enzyme UvrB | |
| 8 | View Details | [1589..1776] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1777..1856] | 2.522879 | Ribosomal protein L32e | |
| 10 | View Details | [1857..1967] | 1.038994 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)