













| General Information: |
|
| Name(s) found: |
SDS23 /
YGL056C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 527 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPQNTRHTSI VEMLSTPPQL PNSTDLNSLS EQTDKNTEAN KSDTESLHKS ISKSSSSSSL 60
61 STLDNTEYSN NNGNSLSTLN SQNLLSVHRQ EWQHTPLSNL VEQNKLIFIR GSISVEEAFN 120
121 TLVKHQLTSL PVENFPGDMN CLTFDYNDLN AYLLLVLNRI KVSNDKITSD CQNGKSVPVA 180
181 EIVKLTPKNP FYKLPETENL STVIGILGSG VHRVAITNVE MTQIKGILSQ RRLIKYLWEN 240
241 ARSFPNLKPL LDSSLEELNI GVLNAARDKP TFKQSRVISI QGDEHLIMAL HKMYVERISS 300
301 IAVVDPQGNL IGNISVTDVK HVTRTSQYPL LHNTCRHFVS VILNLRGLET GKDSFPIFHV 360
361 YPTSSLARTF AKLVATKSHR LWIVQPNDNQ PTASSEKSSS PSPSTPPVTT LPSLASSYHS 420
421 NTQSSRMANS PVLKSSDTSN NKINVNINLS GPSPSQPQSP SATMPPPQSP SNCPASPTPA 480
481 HFEKEYRTGK LIGVVSLTDI LSVLARKQTH HKEIDPQMAR KQRGHIG |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
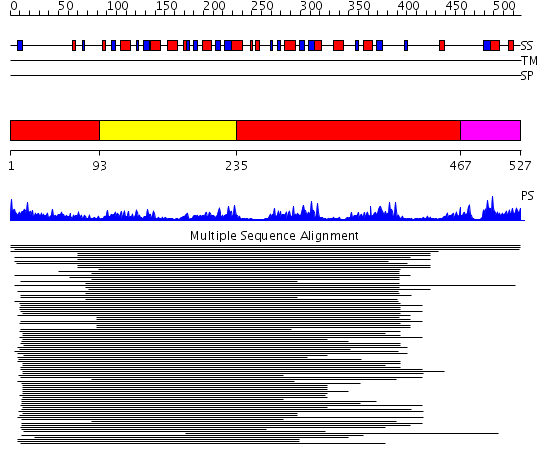
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..92] [235..466] |
41.09691 | Inosine monophosphate dehydrogenase (IMPDH); Type II inosine monophosphate dehydrogenase | |
| 2 | View Details | [93..234] | 41.09691 | Inosine monophosphate dehydrogenase (IMPDH); Type II inosine monophosphate dehydrogenase | |
| 3 | View Details | [467..527] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)