













| General Information: |
|
| Name(s) found: |
HSP78 /
YDR258C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 811 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IDA mitochondrion [IDA |
| Biological Process: |
protein import into mitochondrial matrix
[IDA protein folding [IDA response to stress [IDA mitochondrial genome maintenance [IDA |
| Molecular Function: |
ATPase activity
[ISS unfolded protein binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRQATKAPI QKYLQRTQLL RRSTPRIYTI VQCKRSICSF NARPRVANKL LSDIKTNALN 60
61 EVAISTCALK SSYGLPNFKR TYVQMRMDPN QQPEKPALEQ FGTNLTKLAR DGKLDPVIGR 120
121 DEEIARAIQI LSRRTKNNPC LIGRAGVGKT ALIDGLAQRI VAGEVPDSLK DKDLVALDLG 180
181 SLIAGAKYRG EFEERLKKVL EEIDKANGKV IVFIDEVHML LGLGKTDGSM DASNILKPKL 240
241 ARGLRCISAT TLDEFKIIEK DPALSRRFQP ILLNEPSVSD TISILRGLKE RYEVHHGVRI 300
301 TDTALVSAAV LSNRYITDRF LPDKAIDLVD EACAVLRLQH ESKPDEIQKL DRAIMKIQIE 360
361 LESLKKETDP VSVERREALE KDLEMKNDEL NRLTKIWDAE RAEIESIKNA KANLEQARIE 420
421 LEKCQREGDY TKASELRYSR IPDLEKKVAL SEKSKDGDKV NLLHDSVTSD DISKVVAKMT 480
481 GIPTETVMKG DKDRLLYMEN SLKERVVGQD EAIAAISDAV RLQRAGLTSE KRPIASFMFL 540
541 GPTGTGKTEL TKALAEFLFD DESNVIRFDM SEFQEKHTVS RLIGAPPGYV LSESGGQLTE 600
601 AVRRKPYAVV LFDEFEKAHP DVSKLLLQVL DEGKLTDSLG HHVDFRNTII VMTSNIGQDI 660
661 LLNDTKLGDD GKIDTATKNK VIEAMKRSYP PEFINRIDDI LVFNRLSKKV LRSIVDIRIA 720
721 EIQDRLAEKR MKIDLTDEAK DWLTDKGYDQ LYGARPLNRL IHRQILNSMA TFLLKGQIRN 780
781 GETVRVVVKD TKLVVLPNHE EGEVVEEEAE K |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
HSP60 HSP78 MDJ1 MSP1 YME2 ZIM17 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Hazbun TR, et al. (2003) | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
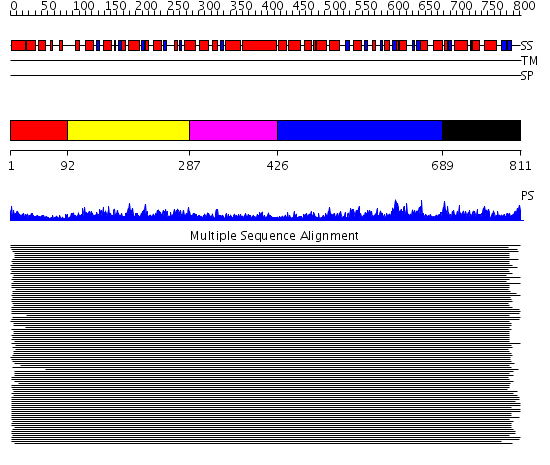
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..91] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [92..286] | 688.124002 | ClpB | |
| 3 | View Details | [287..425] | 8.9 | N-terminal, ClpS-binding domain of ClpA, an Hsp100 chaperone; ClpA, an Hsp100 chaperone, AAA+ modules | |
| 4 | View Details | [426..688] | 7.522879 | Replication factor C | |
| 5 | View Details | [689..811] | 12.61 | HslU |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)