













| General Information: |
|
| Name(s) found: |
MAP2 /
YBL091C
[SGD]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 421 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[TAS |
| Biological Process: |
proteolysis
[IDA |
| Molecular Function: |
methionyl aminopeptidase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTDAEIENSP ASDLKELNLE NEGVEQQDQA KADESDPVES KKKKNKKKKK KKSNVKKIEL 60
61 LFPDGKYPEG AWMDYHQDFN LQRTTVEESR YLKRDLERAE HWNDVRKGAE IHRRVRRAIK 120
121 DRIVPGMKLM DIADMIENTT RKYTGAENLL AMEDPKSQGI GFPTGLSLNH CAAHFTPNAG 180
181 DKTVLKYEDV MKVDYGVQVN GNIIDSAFTV SFDPQYDNLL AAVKDATYTG IKEAGIDVRL 240
241 TDIGEAIQEV MESYEVEING ETYQVKPCRN LCGHSIAPYR IHGGKSVPIV KNGDTTKMEE 300
301 GEHFAIETFG STGRGYVTAG GEVSHYARSA EDHQVMPTLD SAKNLLKTID RNFGTLPFCR 360
361 RYLDRLGQEK YLFALNNLVR HGLVQDYPPL NDIPGSYTAQ FEHTILLHAH KKEVVSKGDD 420
421 Y |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
HKR1 MAP2 |
| View Details | Ho Y, et al. (2002) |
|
MAP2 RFC3 RFC5 RNQ1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
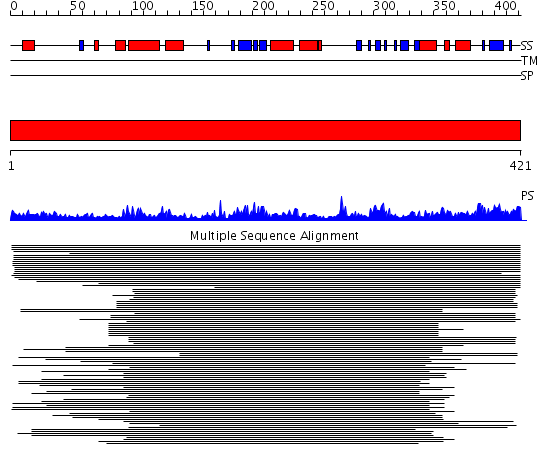
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..421] | 1274.0 | Methionine aminopeptidase, insert domain; Methionine aminopeptidase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)