













| General Information: |
|
| Name(s) found: |
gi|30686918
[NCBI NR]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1072 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
chromatin remodeling complex [IEA] |
| Biological Process: |
chromatin remodeling
[IEA]
ATP-dependent chromatin remodeling [IEA] |
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA] DNA binding [IEA] DNA-dependent ATPase activity [ISS] nucleosome binding [IEA] hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides [IEA] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARASKREVS SDEAYSSEEE EQVNDQANVE EDDDELEAVA RSAGSDEEDV APDEAPVSDD 60
61 EVVPVEDDAE EDEEDEEKAE ISKREKARLK EMQKMKKQKI QQILDSQNAS IDADMNNKGK 120
121 GRIKYLLQQT ELFAHFAKSD PSPSQKKGKG RGRHSSKLTE EEEDEECLKE EEGGIVGSGG 180
181 TRLLTQPACI QGKLRDYQLA GLNWLIRLYE NGINGILADE MGLGKTLQTI SLLAYLHEYR 240
241 GINGPHMVVA PKSTLGNWMN EIRRFCPVLR AVKFLGNPEE RRHIREELLV AGKFDICVTS 300
301 FEMAIKEKTT LRRFSWRYII IDEAHRIKNE NSLLSKTMRL FSTNYRLLIT GTPLQNNLHE 360
361 LWALLNFLLP EVFSSAETFD EWFQISGEND QQEVVQQLHK VLRPFLLRRL KSDVEKGLPP 420
421 KKETILKVGM SQMQKQYYKA LLQKDLEVVN GGGERKRLLN IAMQLRKCCN HPYLFQGAEP 480
481 GPPYTTGDHL VTNAGKMVLL DKLLPKLKDR DSRVLIFSQM TRLLDILEDY LMYRGYQYCR 540
541 IDGNTGGDER DASIEAYNKP GSEKFVFLLS TRAGGLGINL ATADVVILYD SDWNPQVDLQ 600
601 AQDRAHRIGQ KKEVQVFRFC TENAIEAKVI ERAYKKLALD ALVIQQGRLA EQKTVNKDEL 660
661 LQMVRYGAEM VFSSKDSTIT DEDIDRIIAK GEEATAELDA KMKKFTEDAI QFKMDDSADF 720
721 YDFDDDNKDE SKVDFKKIVS ENWNDPPKRE RKRNYSEVEY FKQTLRQGAP AKPKEPRIPR 780
781 MPQLHDFQFF NIQRLTELYE KEVRYLMQAH QKTQMKDTIE VDEPEEVGDP LTAEEVEEKE 840
841 LLLEEGFSTW SRRDFNAFIR ACEKYGRNDI KSIASEMEGK TEEEVERYAQ VFQVRYKELN 900
901 DYDRIIKNIE RGEARISRKD EIMKAIGKKL DRYRNPWLEL KIQYGQNKGK LYNEECDRFM 960
961 ICMVHKLGYG NWDELKAAFR TSPLFRFDWF VKSRTTQELA RRCDTLIRLI EKENQEFDER1020
1021 ERQARKEKKL SKSATPSKRP SGRQANESPS SLLKKRKQLS MDDYVSSGKR RK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
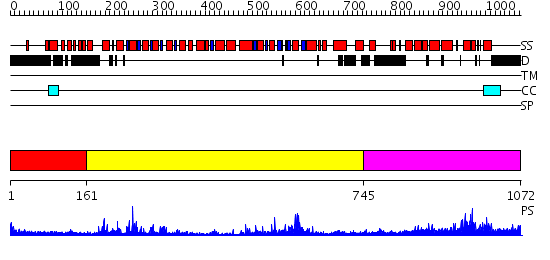
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..160] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [161..744] | 76.09691 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | |
| 3 | View Details | [745..1072] | 54.69897 | nucleosome recognition module of ISWI ATPase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)