













| General Information: |
|
| Name(s) found: |
VHS3 /
YOR054C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 674 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
G1/S transition of mitotic cell cycle
[IGI coenzyme A biosynthetic process [ISS cellular monovalent inorganic cation homeostasis [IGI |
| Molecular Function: |
phosphopantothenoylcysteine decarboxylase activity
[ISS phosphoprotein phosphatase inhibitor activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTNKSSLKNN RKGVASNTLS GAEQANIGSS AMPDTNSTGP FSSVSSLDTP VVRKSTSPTG 60
61 SQTKSIMNAS GTSGAVVSNT PEPGLKRIPT VTFSDPKLGS LRSDVEQTPP NQVARQSSEK 120
121 KATSVHIAAE GANQGRNLKD INTKVPKDGE ASASSFSTPT SILSNADMGN NISSLLAKKL 180
181 SFTGGTDSIL NSDNSSDSPR KEHPHFYVED PLHTPSVRSR SNSTSPRPSV VVNTFNPINI 240
241 EREGSISKTG EPTLLESVLE EAMSPNAVSN PLKRENIMTN MDPRLPQDDG KLHVLFGATG 300
301 SLSVFKLKHM IRKLEEIYGR DKICIQVILT NSATKFFAMK YMRKNKKQHN SIDTSFNSTN 360
361 SNAGNITGNK KKVASLEKFS IQKTSSNSAA SQTNNKQEEE KQMASTTGFP STLGGSRTYS 420
421 NSSNVVSQHP QIELPAHIQF WTDQDEWDVW RQRTDPVLHI ELRRWADILV VAPLTANTLA 480
481 KIALGLCDNL LTSVIRAWNP TFPIFLAPSM GSGTFNSIMT KKHFRIIQEE MPWVTVFKPS 540
541 EKVMGINGDI GLSGMMDANE IVGKIVVKLG GYPDVSAGKE EEEDEDNDEE DDNKKNDTGG 600
601 KDEDNDDDDD DDDDDDDDDD DDDDDDDDDD DDDDDDDDDD DDDDDDDDDD EDDEDEDEDD 660
661 EGKKKEDKGG LQRS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
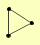
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CAB3 SIS2 VHS3 |
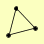
| View Details | Hazbun TR, et al. (2003) |
|
CAB3 SIS2 VHS3 |
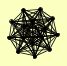
| View Details | Krogan NJ, et al. (2006) |
|
CAB3 CHD1 CKA1 CKA2 CKB1 CKB2 ELF1 HOT1 MSC3 NRG1 PGA2 REH1 RIM13 VHS3 YLR407W |
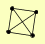
| View Details | Ho Y, et al. (2002) |
|
GLC8 PPZ2 SDS22 VHS3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
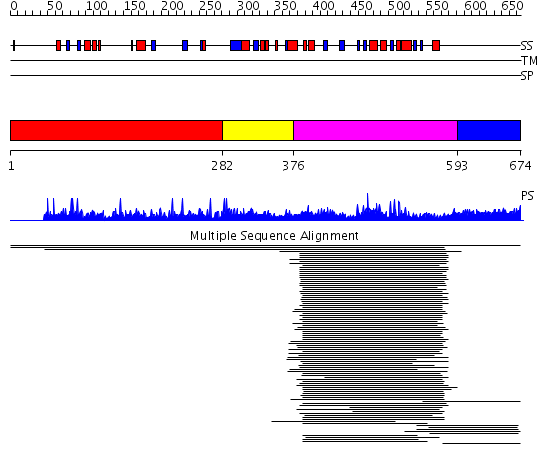
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..281] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [282..375] | 1.88 | 4'-phosphopantothenoylcysteine decarboxylase (PPC decarboxylase, halotolerance protein Hal3a) | |
| 3 | View Details | [376..592] | 371.9897 | 4'-phosphopantothenoylcysteine decarboxylase (PPC decarboxylase, halotolerance protein Hal3a) | |
| 4 | View Details | [593..674] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [574..674] | 4.69897 | Strucuture of the C-terminal domain of human thrombospondin-2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.71 |
Source: Reynolds et al. (2008)