













| General Information: |
|
| Name(s) found: |
ERT1 /
YBR239C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 529 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCTPDENDYK TSTDPDTSAN TNHTLEKKKR KKRKNTNVAC VNCSRLHVSC EAKRPCLRCI 60
61 SKGLTATCVD APRKKSKYLA GIPNRELPMN IQPDLPPRKI MIPIYNNSSN SSLNVNNMGE 120
121 QQKFTSPQHI VHKAKFLSNA ADSEYSILSN IIYQDTLSNK IPIDILYSNT NSTSNSTIGN 180
181 SSNNSPTGTN TSPEETEMEK IRQLYSEQRA NIPPHPYPSS NQNVYSILLG PNSAKIVASQ 240
241 VNLFANHFPL VPVDSADNSL NFKRLLPRDP SEKSSQINWD SSINQYYLNS ETVTFPELAI 300
301 PLKRRKNHLV SVSLESCSPD AANIKSNVEW EHSLRYSTPM EIYTSINAPF SHTPGFHHLL 360
361 VYLKHRFNQQ DLVKMCRSIA EFRPIFIACS VTLTEEDMIF MEQCYQRTLL EYVKFIAQIG 420
421 TPTCIWRRNG QISYVNEEFE ILCGWTREEL LNKMTFIVEI MDDESVRDYF KTLSKVAYRD 480
481 FRGSEKMKVC RLLSPIKGKI IHCCCMWTLK RDVSGLPLMI LGNFMPILN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
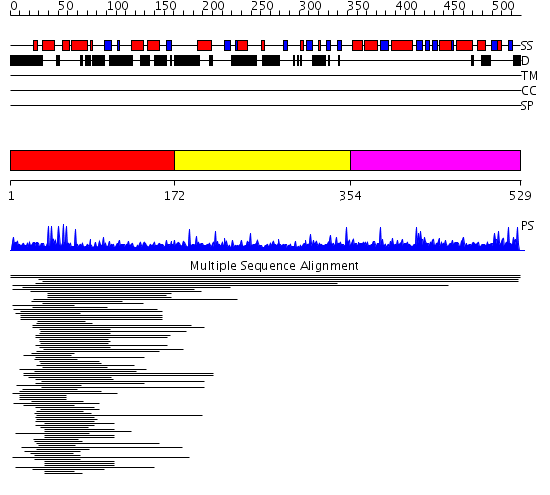
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 9.0 | Hap1 (Cyp1); HAP1 | |
| 2 | View Details | [172..353] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [354..529] | 13.346787 | PAS fold No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)