













| General Information: |
|
| Name(s) found: |
srp-PA /
FBpp0082669
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1264 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[NAS]
|
| Biological Process: |
dorsal closure
[TAS]
larval lymph gland hemopoiesis [TAS] germ-band shortening [IMP][TAS] midgut development [TAS] embryonic hemopoiesis [IMP][TAS] hemopoiesis [IEP][IMP][TAS] endoderm development [TAS] regulation of transcription [NAS] salivary gland morphogenesis [IMP] phagocytosis, engulfment [IMP] embryonic development via the syncytial blastoderm [NAS] positive regulation of transcription, DNA-dependent [IDA] lymph gland development [IMP] endoderm formation [TAS] cell fate determination [TAS] head involution [NAS] regulation of transcription, DNA-dependent [IEA][ISS] amnioserosa maintenance [IMP][TAS] germ cell migration [IMP][TAS] embryonic heart tube development [IMP] defense response [NAS] fat body development [IEP][TAS] crystal cell differentiation [TAS] cellular process [IMP] hemocyte development [IMP][NAS][TAS] mesodermal cell fate commitment [IMP] negative regulation of crystal cell differentiation [TAS] cell fate commitment [TAS] |
| Molecular Function: |
DNA binding
[IDA][TAS]
zinc ion binding [IEA] transcription factor activity [IEA] protein binding [IPI] RNA polymerase II transcription factor activity [ISS] general RNA polymerase II transcription factor activity [NAS] sequence-specific DNA binding [IDA] transcription activator activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTKTTKPKEK AAAGGAVIGS GSGLGSVTKA GGGSLLSNAA DSKIRTAKSN NNKRQAGRAA 60
61 TALAATTTAS ALAATTTAGA TGSNAAANET EIAIETENGE AATPTAAATA AAANLSSLES 120
121 ARSQALTSVV SETARQAVTT ANASATSTST VTAATEIATA TASDTAATSE AAIDDDPSAI 180
181 NTNNNNNNSK AQNDASESVK TKVISYHQSE DQQQQQQQQA QIYEQQQQFL SQQLISHHQQ 240
241 EQHQQAQQQQ HQQVVQEQHQ ASWLAYDLTS GSAAAAAAAA AAASHPHLFG QFSYPPSHHT 300
301 PTQLYEHYPS TDPIMRNNFA FYSVYTGGGG GVGVGMTSHE HLAAAAAAAA AVAQGTTPNI 360
361 DEVIQDTLKD ECFEDGHSTD YHVLTSVSDM HTLKDSSPYA LTHEQLHQQQ HHHQQQLHHH 420
421 QQQQQQLYHQ QQQQQQQQQH HHHHNNSTSS AGGDSPSSSH ALSTLQSFTQ LTSATQRDSL 480
481 SPENDAYFAA AQLGSSLQNS SVYAGSLLTQ TANGIQYGMQ SPNQTQAHLQ QQHHQQQQQQ 540
541 HQQHQQQQLQ QQQQQHHHNQ HQHHNSSSSS PGPAGLHHSS SSAATAAAVA AATAAVNGHN 600
601 SSLEDGYGSP RSSHSGGGGG GTLPAFQRIA YPNSGSVERY APITNYRGQN DTWFDPLSYA 660
661 TSSSGQAQLG VGVGAGVVSN VIRNGRAISA ANAAAAAAAD GTTGRVDPGT FLSASASLSA 720
721 MAAESGGDFY KPNSFNVGGG GRSKANTSGA ASSYSCPGSN ATSAATSAVA SGTAATAATT 780
781 LDEHVSRANS RRLSASKRAG LSCSNCHTTH TSLWRRNPAG EPVCNACGLY YKLHSVPRPL 840
841 TMKKDTIQKR KRKPKGTKSE KSKSKSKNAL NAIMESGSLV TNCHNVGVVL DSSQMDVNDD 900
901 MKPQLDLKPY NSYSSQPQQQ LPQYQQQQQL LMADQHSSAA SSPHSMGSTS LSPSAMSHQH 960
961 QTHPHQQQQQ QLCSGLDMSP NSNYQMSPLN MQQHQQQQSC SMQHSPSTPT SIFNTPSPTH1020
1021 QLHNNNNNNN NSSIFNNNNN NNSSSNENNN KLIQKYLQAQ QLSSSSNSGS TSDHQLLAQL1080
1081 LPNSITAAAA AAAAAAAAAI KTEALSLTSQ ANCSTASAGL MVTSTPTTAS STLSSLSHSN1140
1141 IISLQNPYHQ AGMTLCKPTR PSPPYYLTPE EDEQPALIKM EEMDQSQQQQ QQQQHQQQQH1200
1201 GEIMLSRSAS LDEHYELAAF QRHQQQQQQL QQQTAALLGQ HEQHVTNYAM HKFGVDRETV1260
1261 VKME |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
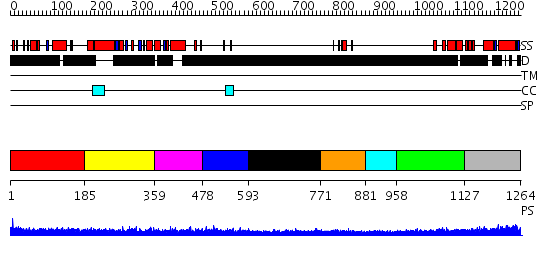
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..184] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [185..358] | 1.115987 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [359..477] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [478..592] | 1.150984 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [593..770] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [771..880] | 5.69897 | Erythroid transcription factor GATA-1 | |
| 7 | View Details | [881..957] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [958..1126] | 1.14198 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1127..1264] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. | ||||||
| 6 |
|
||||||
| 7 | No functions predicted. | ||||||
| 8 | No functions predicted. | ||||||
| 9 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|