













| General Information: |
|
| Name(s) found: |
sl-PA /
FBpp0074009
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1236 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
intracellular signaling pathway
[IEA]
phospholipid catabolic process [IEA] border follicle cell migration [IGI] |
| Molecular Function: |
phosphoinositide phospholipase C activity
[IEA][NAS]
protein binding [IEA] signal transducer activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSCFSAMSAP LLGEMEQTIG MLERGTIVTK LYGKQRRPDR RHLMLIRETR QLLWATVATQ 60
61 TPRTDYEGAI QLREIREIRV GKHSKEFRLF ADDCQRFESS KCFVILHGNH FKLKSFSVVA 120
121 LSEIEADNWV RGLRYMVKDT LGAPYPLQID RWLRREYYQI ENVNTHSAKA TEQSPAQVTI 180
181 KDFKLFLAGV SCKMTTGKFM EHFTEDVRRK HDLKFDDFSR LYQKLLLPNG FASVLSGSGV 240
241 ANFPYSEDQQ VVRPAELKQF LETEQRDVSA SEISSAAIAS FIRDFVQDVE RDVQEPYLTF 300
301 PEFVDFLFSK QNDLWNSKYD QVFMDMNLPL SSYWIASSHN TYLTGDQFSS ESSCEAYARA 360
361 LRMGCRCIEL DCWNGPDNLP YIFHGHTITS KIKFMDVIKT IKDHAFTSSE YPVILSIEQN 420
421 CSLEQQRNMA QALIEVFGDM LLTQPCDRNE QHLPSPYQLR RKIILKHKKL PQFDDIANGI 480
481 SSTGSLGHRS SLGGAGGGAH GENDGENVRK VFKEGLLYFK DPVDKSWNLY QFVLTHQELI 540
541 YSSEINESRN GNSEDDDFGL SSSCSLNSNM QQKQKDTSAN DELHFGENWF HGKLEGGRKE 600
601 ADDLLKKYKH FGDGTFLVRE SATFVGDYSL SFWRRNRPNH CRIKLKHENG SIKYYLVENF 660
661 VFDSLYSLIV YYRKNMLRSS EFSIILKEPV PQPKKHEDQE WFHPNTTKEQ AEQGLYRLEI 720
721 GSFLVRPSVQ SINAFVISFT INRKIKHCRI MQEGRLYGID TMNFESLVSL INYYTRNPLY 780
781 RNVKLSHPVS QELLRQALAE AAQGDHSGGH DDNGASNYMG SNLEENVTCK ALYSYKANKP 840
841 DELSFPKHAI ITNVQRDNSM WWIGDYGGMI KKHLPANYVK VIDSTTEDYN SLNEEGTDGR 900
901 TDSIEIFGAV ASLFESNDPG IIFKLQIQTP TMQNPFVIGF DNQETAYEWI KAIQEAALIA 960
961 SQLASERRKK ERTARVAKEM SDLIIYFRSV PFREHSWIFQ EMSSFPETKA EKQFFQQNTQ1020
1021 LFLSYHRNQI SRVYPKGQRL DSSNFNPMPF WNVGSQMIAL NYQTGDKAMQ LNQAKFRNNG1080
1081 QCGYILKPSF MKSDSFNPNN PLCDGLSEVK VSIRLIAARH LFRGGKSNNP QIVVELIGAS1140
1141 FDTGVKYRTK VNENGFNPVW NESCEFNVRN PQFAILRFEV QDEDMFAETH FIAQACYPLT1200
1201 CIRQGYRSVI LRNKFSEELE LSSLLINIKI ANVTAP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
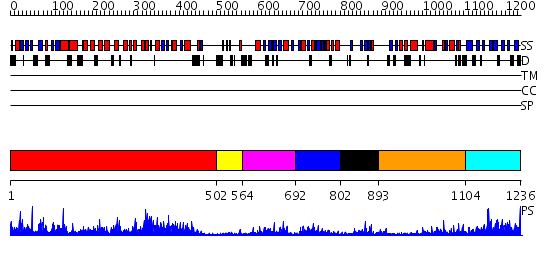
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..501] | 113.0 | No description for 2fjuB was found. | |
| 2 | View Details | [502..563] | 93.69897 | Phosphoinositide-specific phospholipase C, isozyme D1 (PLC-D!); PI-specific phospholipase C isozyme D1 (PLC-D1), C-terminal domain; Phospholipase C isozyme D1 (PLC-D1) | |
| 3 | View Details | [564..691] | 14.0 | No description for 2dx0A was found. | |
| 4 | View Details | [692..801] | 8.154902 | Structural basis for the requirement of two phosphotyrosines in signaling mediated by Syk tyrosine kinase | |
| 5 | View Details | [802..892] | 6.30103 | SOLUTION STRUCTURE OF THE SH3 DOMAIN OF PHOSPHOLIPASE CGAMMA | |
| 6 | View Details | [893..1103] | 4.69 | No description for 2fjuB was found. | |
| 7 | View Details | [1104..1236] | 22.221849 | Synaptogamin I |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)