













| General Information: |
|
| Name(s) found: |
RH51_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 568 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IDA]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA] ATP-dependent helicase activity [IEA][ISS] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVESDKSSVE ELKKRVRKRS RGKKNEQQKA EEKTHTVEEN ADETQKKSEK KVKKVRGKIE 60
61 EEEEKVEAME DGEDEKNIVI VGKGIMTNVT FDSLDLSEQT SIAIKEMGFQ YMTQIQAGSI 120
121 QPLLEGKDVL GAARTGSGKT LAFLIPAVEL LFKERFSPRN GTGVIVICPT RELAIQTKNV 180
181 AEELLKHHSQ TVSMVIGGNN RRSEAQRIAS GSNLVIATPG RLLDHLQNTK AFIYKHLKCL 240
241 VIDEADRILE ENFEEDMNKI LKILPKTRQT ALFSATQTSK VKDLARVSLT SPVHVDVDDG 300
301 RRKVTNEGLE QGYCVVPSKQ RLILLISFLK KNLNKKIMVF FSTCKSVQFH TEIMKISDVD 360
361 VSDIHGGMDQ NRRTKTFFDF MKAKKGILLC TDVAARGLDI PSVDWIIQYD PPDKPTEYIH 420
421 RVGRTARGEG AKGKALLVLI PEELQFIRYL KAAKVPVKEL EFNEKRLSNV QSALEKCVAK 480
481 DYNLNKLAKD AYRAYLSAYN SHSLKDIFNV HRLDLLAVAE SFCFSSPPKV NLNIESGAGK 540
541 VRKARKQQGR NGFSPYSPYG KSTPTKEA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
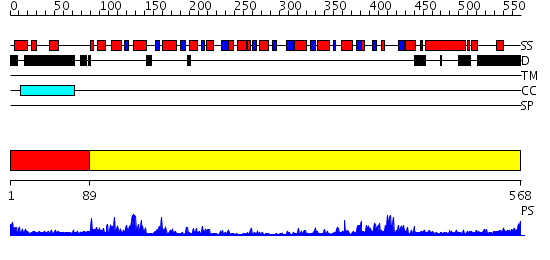
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [89..568] | 29.045757 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)