













| General Information: |
|
| Name(s) found: |
MCM7_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 716 amino acids |
Gene Ontology: |
|
| Cellular Component: |
[NOT]nucleus
[IDA]
cytoplasm [IDA] |
| Biological Process: |
DNA replication initiation
[ISS]
sugar mediated signaling pathway [TAS] DNA unwinding involved in replication [TAS] |
| Molecular Function: |
ATP binding
[ISS]
DNA binding [ISS] DNA-dependent ATPase activity [ISS] protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKDHDFDGDK GLAKGFLENF ADANGRSKYM EILQEVSNRK IRAIQVDLDD LFNYKDESEE 60
61 FLGRLTENTR RYVSIFSAAV DELLPEPTEA FPDDDHDILM TQRADDGTDN PDVSDPHQQI 120
121 PSEIKRYYEV YFKAPSKGRP STIREVKASH IGQLVRISGI VTRCSDVKPL MAVAVYTCED 180
181 CGHEIYQEVT SRVFMPLFKC PSSRCRLNSK AGNPILQLRA SKFLKFQEAK MQELAEHVPK 240
241 GHIPRSMTVH LRGELTRKVS PGDVVEFSGI FLPIPYTGFK ALRAGLVADT YLEATSVTHF 300
301 KKKYEEYEFQ KDEEEQIARL AEDGDIYNKL SRSLAPEIYG HEDIKKALLL LLVGAPHRQL 360
361 KDGMKIRGDV HICLMGDPGV AKSQLLKHII NVAPRGVYTT GKGSSGVGLT AAVMRDQVTN 420
421 EMVLEGGALV LADMGICAID EFDKMDESDR TAIHEVMEQQ TVSIAKAGIT TSLNARTAVL 480
481 AAANPAWGRY DLRRTPAENI NLPPALLSRF DLLWLILDRA DMDSDLELAK HVLHVHQTEE 540
541 SPALGFEPLE PNILRAYISA ARRLSPYVPA ELEEYIATAY SSIRQEEAKS NTPHSYTTVR 600
601 TLLSILRISA ALARLRFSES VAQSDVDEAL RLMQMSKISL YADDRQKAGL DAISDTYSII 660
661 RDEAARSKKT HVSYANALNW ISRKGYSEAQ LKECLEEYAA LNVWQIDPHT FDIRFI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
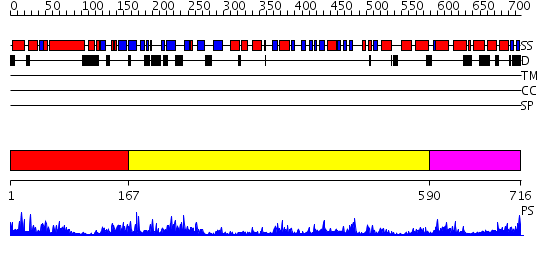
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..166] | 40.09691 | DNA replication initiator (cdc21/cdc54) N-terminal domain | |
| 2 | View Details | [167..589] | 21.30103 | N-terminal, ClpS-binding domain of ClpA, an Hsp100 chaperone; ClpA, an Hsp100 chaperone, AAA+ modules | |
| 3 | View Details | [590..716] | 42.221849 | No description for 2r44A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)