













| General Information: |
|
| Name(s) found: |
KPK1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 465 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
|
| Biological Process: |
response to cold
[IEP]
positive regulation of translation [TAS] response to salt stress [IEP] response to osmotic stress [IDA][IMP] protein amino acid phosphorylation [IDA] |
| Molecular Function: |
protein binding
[IPI]
protein kinase activity [ISS] protein serine/threonine kinase activity [IDA] kinase activity [IDA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVSSQRPVPN KIQKQQYLSI SPSNSVLKDD VELEFSDVFG PLPEEANDIA YDEPAVVYSR 60
61 SHSLVGPCSL DSHSLKLTKL TLLETEDSID LVECLEGESL KENDDFSGND DSDNEKALEG 120
121 DLVKVSGVVG IDDFEVMKVV GKGAFGKVYQ VRKKETSEIY AMKVMRKDHI MEKNHAEYMK 180
181 AERDILTKID HPFIVQLKYS FQTKYRLYLV LDFINGGHLF FQLYHQGLFR EDLARVYTAE 240
241 IVSAVSHLHE KGIMHRDLKP ENILMDTDGH VMLTDFGLAK EFEENTRSNS MCGTTEYMAP 300
301 EIVRGKGHDK AADWWSVGIL LYEMLTGKPP FLGSKGKIQQ KIVKDKIKLP QFLSNEAHAI 360
361 LKGLLQKEPE RRLGSGLSGA EEIKQHKWFK GINWKKLEAR EVMPSFKPEV SGRQCIANFD 420
421 KCWTDMSVLD SPASSPSSDP KANPFTNFTY VRPPPSFLHQ STTTL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
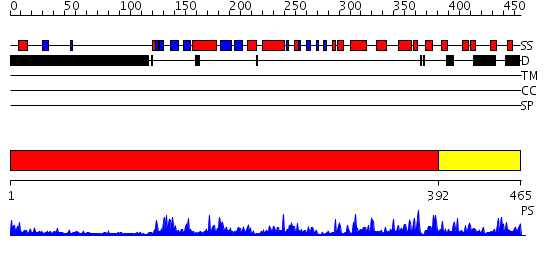
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..391] | 55.0 | CHICKEN SRC TYROSINE KINASE | |
| 2 | View Details | [392..465] | 101.0 | No description for 2jdoA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)