













| General Information: |
|
| Name(s) found: |
set-6 /
CE31454
[WormBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 708 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
chromatin modification
[IEA |
| Molecular Function: |
histone-lysine N-methyltransferase activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MERSRTGGSS TYSMSTANVP IITISDDDDE LTIWEEPRKS PISSNSYSDE RNHSLSSTSN 60
61 SSVERSIIIT ISDDESEATE RNQQEVVDRA ISNNLTEKRR RSVESFHRLH SKERKLEVKS 120
121 RKKSRTSSSS MEASTSCTFP ARQTKTKRHK RKTYSTISSF APHHSTICAE LRNVNPARIA 180
181 DFRHPSIFEY SDMSLNERRE HWKKEMKVIK KHNDIRLMHE EAVRFDDIIE LDPTLNGYYR 240
241 RFMGAEQSTR ALFMAVRCLA TFGRNFYEEP ERDHGEEEIP EELDRYFEKL IYVAPRTRIR 300
301 ITDDIHLSND THRIPVYTDA KGETLKTVKI PNPLLYDHII NNMVDKVKEP LLYEAIKIAG 360
361 SSENVIRCKC CSQDPVVNCF DNKDCPCFIA NQFLQSRNKT TDTNEKLKFH TFQPLMYNDG 420
421 NPTYYNTVGF ACSPKCACKG ACTNNATYLI QKKLYSIEIY RADPQIGFGI RSTLFIPAGT 480
481 PIIEYCGELV DGERLHSSLE NYSYQLTDCE GDKHLYNLLR EKYKNNPEYY DVLDELSKHH 540
541 FHLDAKMQGS VGRFANHSCT PNMEPLRLFK EGFTPANMRM IFFTLKDIFP GEPLTLDYGS 600
601 EYKVFRRQRC LCRTFACRSG PHYEKFSNLD GKLIASCFVR LHHASRIRYS VDLQLIERFY 660
661 SVGKVEEVRV DPCVSFSKYN LKWRRQLSTA TCERLENVIE IELSDEDS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
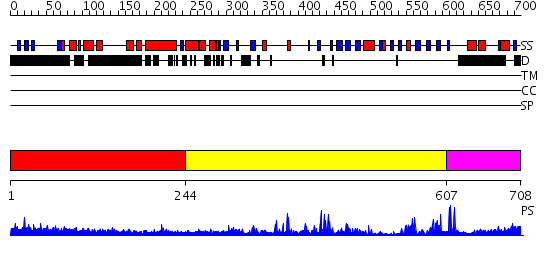
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..243] | 13.69897 | Initiation factor eIF2 gamma subunit, domain II; Initiation factor eIF2 gamma subunit; Initiation factor eIF2 gamma subunit, N-terminal (G) domain | |
| 2 | View Details | [244..606] | 51.045757 | No description for 2igqA was found. | |
| 3 | View Details | [607..708] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)