













| General Information: |
|
| Name(s) found: |
CE23016
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 773 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
protein catabolic process
[IEA proteolysis [IEA ATP-dependent proteolysis [IEA |
| Molecular Function: |
nucleotide binding
[IEA ATPase activity [IEA ATP binding [IEA serine-type endopeptidase activity [IEA nucleoside-triphosphatase activity [IEA ATP-dependent peptidase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKIEENMELP VILVTSGVLL PGASLKIPIR SKLNIQTIEK YLTRSSNDNY VVIAYKVSTD 60
61 KVYEVATIAY VEKLFGWTFN STVHYSLDVI GLHRANIDKL SLPTCIVSKV VDLNEAISNQ 120
121 NAIEKLVTGA KIIASNSLTD KFSREIYSLI DEKEYGKLAD LCVSQMKFLG FMQLLEFLGA 180
181 NGTDARVEMC IKWMNEKKDA NTLKLKVPNS LEASFPVDGK KRKIPNVKNQ VEQLEEKLNA 240
241 IEFSDEVSDR VYSELHRLKS MNAQQSEYNI LMNWLELVSS LPWNTSTIDD IELHKARTIL 300
301 TESHEAMDDV KERVLEHLAV CKMNNSVKGM ILCFTGPPGI GKTSIAKAIA ESMGRKFQRV 360
361 SLGGIRDESD IRGHRRTYVA AMPGRIIEAL KTCKTNNPVF LLDEVDKLYS GNQGSPSAAL 420
421 LELLDPEQNS TFHDHYLNIP FDVSKIMFIA TANDIDRLEP ALRDRLEIIE MSGYSLKEKV 480
481 KICENHLLTR QLTKHCISHD YVKLERQAIV AMIEEYTMEA GVRQLERNVG AICRNVALRL 540
541 AEALNSDPGA DVLPVMELPI QISASNIHKI LKNKHMKRVK IVEKMRPLPA GVCFGLSVTT 600
601 IGGRVMPIEA SKSKGTGKIV TTGHLGKVLK ESILVAKGWL SANSERLGLG TLEDQDIHVH 660
661 LPAGAVNKDG PSAGTGLACA LVSLATNIPL RSDAAVTGEI SLTGHVLPIG GVKEKVLAAQ 720
721 REGLRRVVLP KSNEEEYLKM DEDIRLEMDV VLAETIEDVI GAMMDKSPVL AKL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
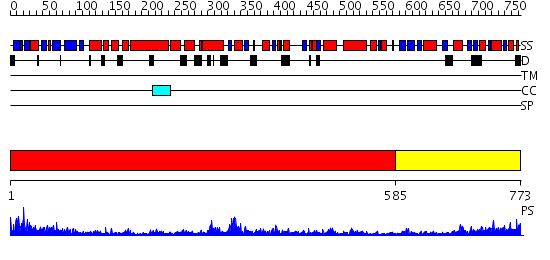
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..584] | 74.69897 | Crystal Structure Analysis of ClpB | |
| 2 | View Details | [585..773] | 39.39794 | Catalytic domain of E.coli Lon protease |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.45 |
Source: Reynolds et al. (2008)