













| General Information: |
|
| Name(s) found: |
adm-4 /
CE15265
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 686 amino acids |
Gene Ontology: |
|
| Cellular Component: |
integral to membrane
[IEA |
| Biological Process: |
proteolysis
[IEA reproduction [IMP |
| Molecular Function: |
zinc ion binding
[IEA metalloendopeptidase activity [IEA metallopeptidase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKIQDRSLLI FLVLGILKSD AFNTRVKRHA PIRFQRSTRQ SVVHFEFLDQ EYVVDLEPNH 60
61 STFHENFKVF TQDGPQIVPR DEYIGTVREP RAGRAVLTQL EENVYIGSLY FVDDTLHLEP 120
121 SYPHQLSDDL GPVVGYFESD LDLNLDLSAM PVRNQVSFRR ANPFLKHRRA IAIPSDRRKD 180
181 VLNVKRNRCT LKLVADYSFY SIFGKNNTGI VTKFLVNMIA RVNEIYTPIN WDVGKEDDIS 240
241 GRGRFQNMGF SIKEIKVLDR PNASDSHYNS YSRIWEVERL LREFAFAEGS KDFCLVHLVT 300
301 ARTFREVATL GLAYVSYKKW DETAGGICSK QETFNGRVAY INVLLSTSFA NSEQSTYPLI 360
361 TKEIDIVVSH EYGHAWGATH DPTIDSDDPD VEECSPNDQN GGKYLMSQYA QKGYDANNVL 420
421 FSPCSRKLIR DVLIGKWESC FQEEMTSFCG NGIVEDGEEC DNGVDTDNEF NCCDKFCRLA 480
481 VGAKCSPLNH ICCTPTCQFH NSTHVCLPGD SLLCKADAVC NGFSGECPSA PPVRDGQECL 540
541 EGGECLNGVC LPFCEKMSIG KKSCICEDLE LSCRLCCRDY NGTCAPVPGH VYLRDGVRCS 600
601 KGSCRDRKCV NEVVDNVRNY FLITFQTTGG VLEFIKTHIV VIAIIFFTLI FVGIYKIVKY 660
661 GENFTEKVTH KTAGGCRSVF VKADVN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
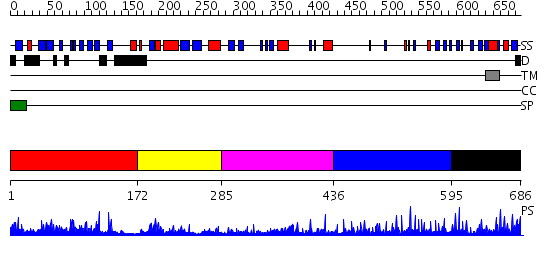
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..171] | 1.195987 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [172..284] | 30.0 | No description for 2i47A was found. | |
| 3 | View Details | [285..435] | 5.89 | Crystal structure of the catalytic domain of human ADAM33 | |
| 4 | View Details | [436..594] | 3.0 | No description for 2ao7A was found. | |
| 5 | View Details | [595..686] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|