













| General Information: |
|
| Name(s) found: |
CE08773
[WormBase]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 459 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
electron transport
[IEA removal of superoxide radicals [IEA glutathione metabolic process [IEA cell redox homeostasis [IEA detoxification of mercury ion [IEA |
| Molecular Function: |
disulfide oxidoreductase activity
[IEA NADP or NADPH binding [IEA oxidoreductase activity [IEA FAD binding [IEA mercury ion binding [IEA mercury (II) reductase activity [IEA thioredoxin-disulfide reductase activity [IEA glutathione-disulfide reductase activity [IEA dihydrolipoyl dehydrogenase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGVKEFDYL VIGGGSGGIA SARRAREFGV SVGLIESGRL GGTCVNVGCV PKKVMYNCSL 60
61 HAEFIRDHAD YGFDVTLNKF DWKVIKKSRD EYIKRLNGLY ESGLKGSSVE YIRGRATFAE 120
121 DGTVEVNGAK YRGKNTLIAV GGKPTIPNIK GAEHGIDSDG FFDLEDLPSR TVVVGAGYIA 180
181 VEIAGVLANL GSDTHLLIRY DKVLRTFDKM LSDELTADMD EETNPLHLHK NTQVTEVIKG 240
241 DDGLLTIKTT TGVIEKVQTL IWAIGRDPLT KELNLERVGV KTDKSGHIIV DEYQNTSAPG 300
301 ILSVGDDTGK FLLTPVAIAA GRRLSHRLFN GETDNKLTYE NIATVVFSHP LIGTVGLTEA 360
361 EAVEKYGKDE VTLYKSRFNP MLFAVTKHKE KAAMKLVCVG KDEKVVGVHV FGVGSDEMLQ 420
421 GFAVAVTMGA TKKQFDQTVA IHPTSAEELV TMRGGVKPE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
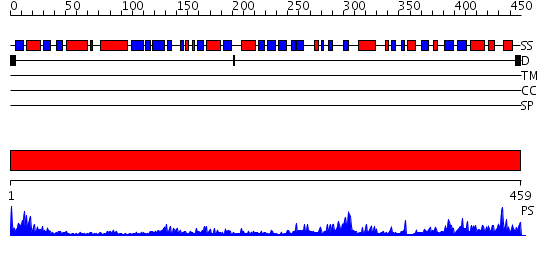
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..459] | 98.69897 | HUMAN GLUTATHIONE REDUCTASE A34E/R37W MUTANT |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)