













| General Information: |
|
| Name(s) found: |
CE05752
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 284 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
estrogen biosynthetic process
[IEA tetrahydrobiopterin biosynthetic process [IEA poly-hydroxybutyrate biosynthetic process [IEA metabolic process [IEA fatty acid biosynthetic process [IEA cellular metabolic process [IEA |
| Molecular Function: |
estradiol 17-beta-dehydrogenase activity
[IEA sepiapterin reductase activity [IEA coenzyme binding [IEA 3-oxoacyl-[acyl-carrier-protein] reductase activity [IEA catalytic activity [IEA oxidoreductase activity [IEA NAD or NADH binding [IEA 3-hydroxybutyrate dehydrogenase activity [IEA acetoacetyl-CoA reductase activity [IEA 2-deoxy-D-gluconate 3-dehydrogenase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRFTDKVAI ITGSSNGIGQ ATARLLASEG AKVTVTGRNA ERLEETKNIL LGAGVPEGNV 60
61 LVVVGDITQE SVQENLIKST LDKFGKIDIL VNNAGAGIPD AQGKSGVNQS IDTYHKTFEL 120
121 NVQSVIEMTQ KARPHLAKTQ GEIVNISSIG AGPAAQVASP YYSIAKAALD QYTRTAAIDL 180
181 VPEGIRVNSV SPGAVSTGFS AVSRGLTEEK SKAFYDYLGA QRECIPRGFC AVPEDIAKVI 240
241 AFLADRNASN YIIGQTIVAD GGTSLILGAH AHGAKMMQAV FKEL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
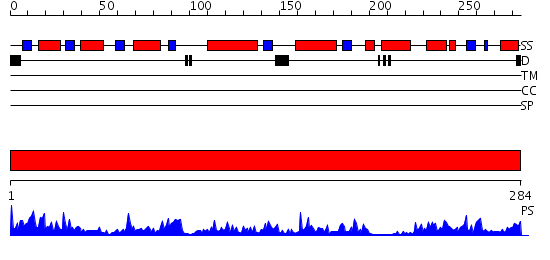
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..284] | 68.045757 | Crystal Structure of Short-Chain Dehydrogenase/Reductase of unknown Function from Caenorhabditis Elegans with Cofactor |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.65 |
Source: Reynolds et al. (2008)