













| General Information: |
|
| Name(s) found: |
SPAP8A3.05
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 695 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA exosome (RNase complex) [ISS] |
| Biological Process: |
mRNA catabolic process
[ISS |
| Molecular Function: |
GTP binding
[ISS GTPase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRLSQLLNS KKAKQKPPSE HPIGLSSILK QDSSSSSDSP NFFPSSSTND HQERDTINDT 60
61 NFVVPEKQKT SKLALLAAER KKLHSSFPST QQQPPKTEKE KEKEPIQAKH KKNVENDFLL 120
121 QRFRKVRIAE KKDSEQPSSH EIHLTDDDDK TTLQKQMVES DQLKKNPQEV KLAPPSSFAK 180
181 CLTGAKKRVF EDQIEIHLSK SSLLGFNAPS PDDIVLMAQS KSKSFQKHKR LDEQLLNSVK 240
241 SMKKVSQQLK PQKNTNDSNN DHTLLSQDQL IELSKLVKPR TKLLLLGPPK SGKKTLLSRL 300
301 FFQIGSFDPK TMQKCTVLNA KKESLSSVLK STKTKWYDFE TFSNSYSSTI IDFPLGIFTT 360
361 NASSRDNFLK HSSLFQVMNT AIFTIDCLNP LEGLDGISSI LQLMNGLSIS SYMFAITKMD 420
421 EIEWDENKFI NLVNSIQSFL KESCGIIEKS KFIPISGLKG TNLTSISQEK LSQWYKSDTL 480
481 LGKIDKEADT NHGTWNFLLN LPLSLTISHI TPLPENQSHI YCSIHSGMLQ DSQKLYVGTG 540
541 RLETQITGLS SDENPKGFNV AGDMIQAKIP TLPNLCPGIL IADSIDAFTS SKTAYVNATW 600
601 FHGSLEKGKS MHVIALFGCH AVLTKLYCFT DSQEKAPNAI GNDLERNRTS LVKIELENAF 660
661 PLVKESYINT LSRVLFVSEK WNSLIAFGTV LSLHD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
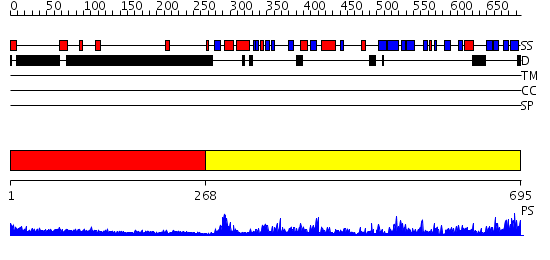
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..267] | 5.0 | Heavy meromyosin subfragment | |
| 2 | View Details | [268..695] | 133.0 | Elongation factor eEF-1alpha, domain 2; Elongation factor eEF-1alpha, C-terminal domain; Elongation factor eEF-1alpha, N-terminal (G) domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)