













| General Information: |
|
| Name(s) found: |
SPBC4F6.17c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 803 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[ISS |
| Biological Process: |
protein import into mitochondrial matrix
[ISS cellular response to stress [ISS protein folding [ISS positive regulation of mitochondrial translation in response to stress [ISS] mitochondrial genome maintenance [ISS |
| Molecular Function: |
ATPase activity
[ISS ATP binding [ISS unfolded protein binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNILPKSIPI RIQSLRRYQG SLKTSLRALS TRPIFEKRLL AETPFSIRPS NASVLIAKRS 60
61 PKFGWKQVRF YAANPNGGNF RMDLGGGRKQ GAALEEYGTD LTALAKQGKL DPVIGREEEI 120
121 QRTIQILSRR TKNNPALVGP AGVGKTAIME GLASRIIRGE VPESMKDKRV IVLDLGALIS 180
181 GAKFRGDFEE RLKSVLSDLE GAEGKVILFV DEMHLLLGFG KAEGSIDASN LLKPALARGK 240
241 LHCCGATTLE EYRKYIEKDA ALARRFQAVM VNEPSVADTI SILRGLKERY EVHHGVRITD 300
301 DALVTAATYS ARYITDRFLP DKAIDLVDEA CSSLRLQQES KPDELRRLDR QIMTIQIELE 360
361 SLRKETDTTS VERREKLESK LTDLKEEQDK LSAAWEEERK LLDSIKKAKT ELEQARIELE 420
421 RTQREGNYAR ASELQYAIIP ELERSVPKEE KTLEEKKPSM VHDSVTSDDI AVVVSRATGI 480
481 PTTNLMRGER DKLLNMEQTI GKKIIGQDEA LKAIADAVRL SRAGLQNTNR PLASFLFLGP 540
541 TGVGKTALTK ALAEFLFDTD KAMIRFDMSE FQEKHTIARL IGSPPGYIGY EESGELTEAV 600
601 RRKPYAVLLF DELEKAHHDI TNLLLQVLDE GFLTDSQGRK VDFRSTLIVM TSNLGSDILV 660
661 ADPSTTVTPK SRDAVMDVVQ KYYPPEFLNR IDDQIVFNKL SEKNLEDIVN VRLDEVQQRL 720
721 NDRRIILTVT EAARKWLAEK GYSPAYGARP LNRLIQKRIL NTMAMKIIQG EIKSDENVVI 780
781 DVLDGELEFK ANKLEPQSTS HED |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Liu X, et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
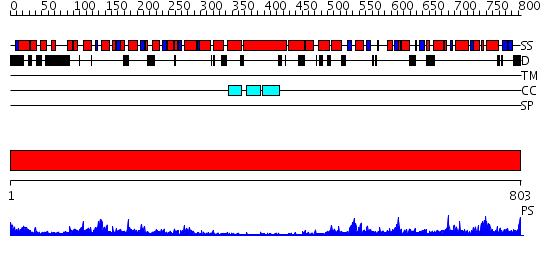
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..803] | 143.0 | Crystal Structure Analysis of ClpB |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)