













| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAILKRTNRA KAATAAAPNS TGKSNGIKKA VYTSTRKKTV GVDDLTLLSK ITDEEINKNL 60
61 ELRFRNGEIY TYIGHVLISV NPFRDLGIYT MDILKSYQGK NRLETSPHVY AIAENAYYQM 120
121 KSYHENQCII ISGESGAGKT EAAKRIMQYI THVSKSVGTE IERVSEIILA TNPLLESFGC 180
181 AKTLRNNNSS RHGKYLEMIF NSGGVPVGAK ITNYLLEKNR IVNQVRNERN FHIFYQFTKS 240
241 APQKYRDTYG IQGPENYVYT SACQCLSVDG ISDEKDFQGT MNAMKVIGIT EPEQDEIFRM 300
301 LSIILWLGNI QFQEGQDGGS VISDKSITEF LGYLIGVPVA AIERALTIRI MQTQHGARRG 360
361 SVYEVPLNPT QALAVRDALS MAIYNCLFDW IVERVNKALV TSDNSVSNSI GILDIYGFEI 420
421 FENNSFEQLC INYVNEKLQQ IFIELTLKTE QEEYVREQIA WTPIKYFNNK VVCDLIESKR 480
481 PPGLFAAMND AIATAHADSA AADSAFAQRL NFLSSNPHFE QRQNQFIVKH YAGDVTYSIT 540
541 GMTDKNKDQL ATDILNLIHS SNNEFMKSIF PVAEESNSRR RPPTAGDRIK TSANDLVETL 600
601 MKCQPSYIRT IKPNQTKSPN DYDQQMVLHQ IKYLGLQENI RIRRAGFAYR QAFDTFAQRF 660
661 AVLSGKTSYA GEYTWQGDDK SACEQILKDT NIPSSEYQMG TSKVFIKNPE TLFALEDMRD 720
721 KFWDTMATRI QRAWRSYVRR RSEAAACIQK LWNRNKVNME LERVRNEGTK LLQGKKQRRR 780
781 YSILGSRKFY GDYLSASKPN GTLWNTCGLS QNDHVIFSMR CEVLVHKLGR TSKPSPRQLV 840
841 LTKKNLYLVI TKIVDQKLTQ QVEKKFAVSS IDSVGLTNLQ DDWVAIRNKS SQNGDMFLRC 900
901 FFKTEFITTL KRINRNIQVI VGPTIQYCRK PGKVQTVKTA KDETTKDYDY YKSGTIHVGT 960
961 GLPPTSKSKP FPRLATGGST AAARGPRPVV QNKPAATKPV SMPAAKSKPA PMANPVSTAQ1020
1021 QTQNRPPAPA MQARPNTTQA AAPVTSTTTT IKQATTVSAS KPAPSTVTSA ASSPSNISKP1080
1081 SAPVANNVSK PSAVPPPPPP PPAEVEKKDL YLALYDFAGR SPNEMTIKKD EIIEIVQKEP1140
1141 SGWWLALKNG AEGWVPATYV TEYKGSTPQT TASSTNVAAQ ANNNASPAEV NNLAGSLADA1200
1201 LRMRASAVRG SDEEEDW |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Liu X, et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
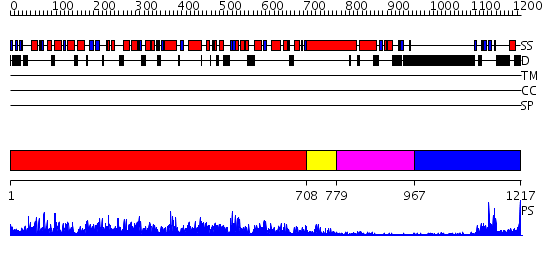
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..707] | 1000.0 | Myosin S1, motor domain | |
| 2 | View Details | [708..778] | 2.07 | No description for 2ix7C was found. | |
| 3 | View Details | [779..966] | 69.69897 | No description for PF06017.4 was found. No confident structure predictions are available. | |
| 4 | View Details | [967..1217] | 14.69897 | No description for 2h8hA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.57 |
Source: Reynolds et al. (2008)