













| General Information: |
|
| Name(s) found: |
fft3 (reserved) /
SPAC25A8.01c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 922 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC nuclear chromatin [IDA chromatin remodeling complex [ISS |
| Biological Process: |
chromatin remodeling
[ISS positive regulation of Ran protein signal transduction [IGI |
| Molecular Function: |
ATP binding
[ISS DNA binding [ISS DNA-dependent ATPase activity [IDA protein binding [IPI helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDGKRKIEHT ADGTHYDATS NVKRKPIFPP FIADSLSEAT EKANVMGGGM NSRLQILSEM 60
61 SKRVQATAPI SSLEHFKQLS DISPSFTSSA NSINQPYNYS GSLENLVPTP SAGTPSQFMD 120
121 AQNPYGAVYN ALSQFSETEP KMPSYMDDEE ASDSLPLSLS SQSLSSQVTN QKPAPHRLTM 180
181 RERYAANNLT NGLQFTLPLS SRKTYEPEAD DDSNDDMYSD DDSNADRWAS RIDTAALKEE 240
241 VLKYMNRCST QDLADMTGCT LAEAEFMVAK RPFPDLESAL VVKQPRPVIP KGRRGRREKT 300
301 PLGPRLVGIC MEIMRGYFVV DALIRQCEQL GGKIQRGIEA WGLSNTATSD EGETSLVNFD 360
361 QMKSFGTPAN SSFITTPPAS FSPDIKLQDY QIIGINWLYL LYELKLAGIL ADEMGLGKTC 420
421 QTIAFFSLLM DKNINGPHLV IAPASTMENW LREFAKFCPK LKIELYYGSQ VEREEIRERI 480
481 NSNKDSYNVM LTTYRLAATS KADRLFLRNQ KFNVCVYDEG HYLKNRASER YRHLMSIPAD 540
541 FRVLLTGTPL QNNLKELISL LAFILPHVFD YGLKSLDVIF TMKKSPESDF ERALLSEQRV 600
601 SRAKMMMAPF VLRRKKSQVL DALPKKTRII EFCEFSEEER RRYDDFASKQ SVNSLLDENV 660
661 MKTNLDTNAN LAKKKSTAGF VLVQLRKLAD HPMLFRIHYK DDILRQMAKA IMNEPQYKKA 720
721 NELYIFEDMQ YMSDIELHNL CCKFPSINSF QLKDEPWMDA TKVRKLKKLL TNAVENGDRV 780
781 VLFSQFTQVL DILQLVMKSL NLKFLRFDGS TQVDFRQDLI DQFYADESIN VFLLSTKAGG 840
841 FGINLACANM VILYDVSFNP FDDLQAEDRA HRVGQKKEVT VYKFVVKDTI EEHIQRLANA 900
901 KIALDATLSG NAETVEAEDD DD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
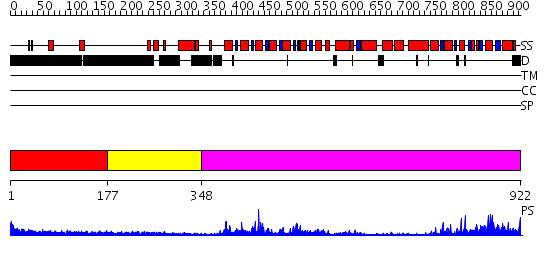
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..176] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [177..347] | 1.68 | Architecture of the photosynthetic oxygen evolving center | |
| 3 | View Details | [348..922] | 77.09691 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)