













| General Information: |
|
| Name(s) found: |
ctf18 /
SPBC902.02c
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 960 amino acids |
Gene Ontology: |
|
| Cellular Component: |
Ctf18 RFC-like complex
[TAS nucleus [IDA nuclear replication fork [IC] cytosol [IDA DNA replication factor C complex [ISS |
| Biological Process: |
attachment of spindle microtubules to kinetochore involved in homologous chromosome segregation
[IMP sister chromatid cohesion [TAS meiotic chromosome segregation [IMP DNA-dependent DNA replication [IC] |
| Molecular Function: |
ATP binding
[IEA]
DNA clamp loader activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSIPNEDDL EKLLVAEQNV PYGKNYESFE DEENYLLELR NANALENLEH RSIPNDLFHS 60
61 SQPVGSPTRN GDDIPSTLDL YSSDNAAIDT DISEDETINQ RHAPQTDYRY PNTSANPKMG 120
121 LEESMDIDMP SIDLDISNAA AFPRDSNLLF RNYNSHTKTS EKGSPVNFEK ENQDSNVLFS 180
181 TGRTSVLHID AEQSQTNNTV TPLKDDALSL FSFENDVNGA VNDNSLDQKL SASKNSQRVS 240
241 LPFFSKIELD EGSSFSGPKI SARTSSGKII YFPKKKNRHS DGLLLQPKRL ADEISQVNEI 300
301 GKDFNQDLLN SVRIWSENFL IVSKEKSVAL KTEKLDSLQI TNCTTKPQSQ KLWVDTYRPQ 360
361 LFRDLLGDER VHRAAMHWIK AWDPCVFGKS RLQPSKSMRF NPRFTNITSD SDRPDKRIMM 420
421 LTGLAGAGKT TLAHVIAHQA GYKVLEINAS DDRTAHTVHE KVSSAISNHS ALSSQPTCVI 480
481 VDEIDGGDPA FVRALLSLLE SDEKATEYSQ AGNSKKKKKF KKLCRPIICI CNDLYTPALR 540
541 PLRPYAQIIY FRPPPQASLV GRLRTICRNE NIAVDSRSLT LLTDIYNSDI RSCINSLQLL 600
601 SLNNKRIDSE TIKLLQPKSN SFSTSSLIQS LFLQLDNKQI RAIEASQPTY SHLDALLARI 660
661 DGANDSESVL MNCFHTYLDL PFTDSLLSKP ALTSEWLYFF DQLHSQCYKG NYELWRYIPY 720
721 SIIHFHYLYA TPEKCRLPHP PRSDLEALKL YRTRKEILDS FISTLNAYEN QMHGERSILL 780
781 ELIRTILITI NPTLKQKEDS MPRSALPSKI EHAINILNHY NLRFQQLPVG DGNYVYRLEP 840
841 PLDELVWDAP TSSYSVRQML SQELLKKRLA DIKKQTLTDN PTSSNSSRKR KDFNSGKAIK 900
901 RDFFGRIISE PKSEAVTSNN AALNTGDHPV SIKHAINIKF HDGFSNAVRK PISLNEILNF 960
961 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
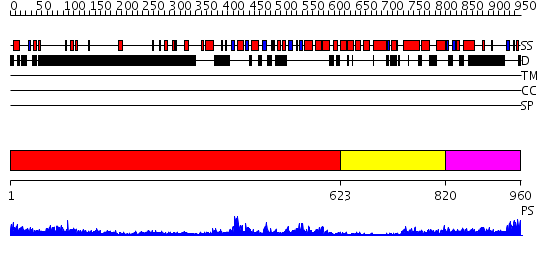
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..622] | 67.30103 | N-terminal, ClpS-binding domain of ClpA, an Hsp100 chaperone; ClpA, an Hsp100 chaperone, AAA+ modules | |
| 2 | View Details | [623..819] | 56.69897 | EDTA treated | |
| 3 | View Details | [820..960] | 84.0 | VCP/p97 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)