













| General Information: |
|
| Name(s) found: |
imp1 /
SPBC1604.08c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 539 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear envelope
[IDA nucleus [IDA cytosol [IDA nucleolus [IDA nuclear pore [TAS |
| Biological Process: |
nucleocytoplasmic transport
[TAS M phase of mitotic cell cycle [IMP establishment of nucleus localization [IMP NLS-bearing substrate import into nucleus [IDA cellular response to stress [IGI regulation of mitotic cell cycle [IMP |
| Molecular Function: |
GTP binding
[NAS protein binding [IPI protein transporter activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESRYLSDRR SRFKSKGVFK ADELRRQREE QQIEIRKQKR EESLNKRRNL NAVLQNDIDV 60
61 EEEADQSQVQ MEQQMKDEFP KLTADVMSDD IELQLGAVTK FRKYLSKETH PPIDQVIACG 120
121 VVDRFVQFLE SEHHLLQFEA AWALTNIASG TTDQTRIVVD SGAVPRFIQL LSSPEKDVRE 180
181 QVVWALGNIA GDSSACRDYV LGNGVLQPLL NILQSSASDV SMLRNATWTL SNLCRGKNPP 240
241 PNWSTISVAV PILAKLLYSE DVEIIVDACW AISYLSDGPN EKIGAILDVG CAPRLVELLS 300
301 SPSVNIQTPA LRSVGNIVTG TDAQTQIIID CGALNAFPSL LSHQKENIRK EACWTISNIT 360
361 AGNTQQIQAI IESNLIPPLV HLLSYADYKT KKEACWAISN ATSGGLGQPD QIRYLVSQGV 420
421 IKPLCDMLNG SDNKIIQVAL DAIENILKVG EMDRTMDLEN INQYAVYVEE AGGMDMIHDL 480
481 QSSGNNDIYL KAYSIIEKYF SDEDAVEDLA PETENGAFTF GNGPQAQGEF KFDSQDMAM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
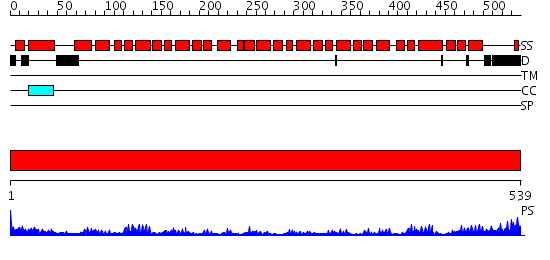
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..539] | 90.522879 | Crystal structure of the exportin Cse1p complexed with its cargo ( Kap60p) and RanGTP |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)