













| General Information: |
|
| Name(s) found: |
brl2 /
SPCC970.10c
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 680 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA HULC complex [IDA centromeric heterochromatin [IDA |
| Biological Process: |
chromatin assembly
[IMP DNA replication-independent nucleosome assembly [TAS histone H2B ubiquitination [IDA histone H3-K4 methylation [IMP regulation of transcription from RNA polymerase II promoter [IEP |
| Molecular Function: |
ubiquitin-protein ligase activity
[IDA zinc ion binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYQNGKPDAP TILGQKRELE DVEIQDDDIQ EVSKEDLLKD VRVRSIQFDE LESKIEGLQN 60
61 LAEEKLKVLA TLVSWWPEIL QQFSVVFQGN ELKDFESEGV FSILEKFPEL SYFNDAVKNN 120
121 KTKALSIIQK LLSTVDSSTN SVSRDPFSVL SIDDSALTEK LNTINLDIDK ILDELDTTRS 180
181 QLHSIIKLPD RSSSFTLQCI NESVRPQSTK VKEEATTSSK GKDEEKKVST VEQRTQLQQL 240
241 SRLQDQQNGL MESRSQSLKI LDSNVNEMDK LIMERENALN NVETTNLKKY SSFLALKEAV 300
301 SMTSEQLRVL EHLLSECSHE INVLSQQSKN FNGVFESSYQ PLINDLDHQI SVMQNDEKRI 360
361 NNAKTELSLS LEKKLEAKKQ KEKVYKDKLD ELANLETMVL EKKKAVATRE AANKIRLVDL 420
421 NDLELQKDLS TYLSKELAST EKAFRLVKQQ TVKSSHSHYQ ELITKFSVEK EKAEQKYFLT 480
481 MKSTDSLHAE VKLLRQKYQK TNEIISKMLN SQDTAVHRII EFEDQLARLS SVRNNSIKQS 540
541 TTFQVKKSSQ KSTIQNLEEK VSYLQQFMDK NNATLTDLEF QCSDLSSSID ILSKQDEEHE 600
601 KEKRKLKDTG VSTSAEELKT FRAMCKCSVC NFERWKDRII SLCGHGFCYQ CIQKRIETRQ 660
661 RRCPICGRGF GASDVIPIHL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
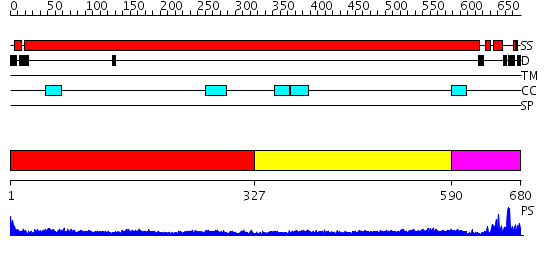
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..326] | 18.30103 | Heavy meromyosin subfragment | |
| 2 | View Details | [327..589] | 10.522879 | Tropomyosin | |
| 3 | View Details | [590..680] | 5.0 | Eea1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)