













| General Information: |
|
| Name(s) found: |
wis1 /
SPBC409.07c
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 605 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA cytosol [IDA |
| Biological Process: |
response to arsenic
[IMP response to osmotic stress [TAS response to hydrogen peroxide [IMP protein amino acid phosphorylation [IGI regulation of translation in response to stress [IMP regulation of mitotic cell cycle [IMP stress-activated MAPK cascade [TAS |
| Molecular Function: |
ATP binding
[ISS protein binding [IPI MAP kinase kinase activity [IGI protein serine/threonine kinase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSPNNQPLS CSLRQLSISP TAPPGDVGTP GSLLSLSSSS SSNTDSSGSS LGSLSLNSNS 60
61 SGSDNDSKVS SPSREIPSDP PLPRAVPTVR LGRSTSSRSR NSLNLDMKDP SEKPRRSLPT 120
121 AAGQNNIGSP PTPPGPFPGG LSTDIQEKLK AFHASRSKSM PEVVNKISSP TTPIVGMGQR 180
181 GSYPLPNSQL AGRLSNSPVK SPNMPESGLA KSLAAARNPL LNRPTSFNRQ TRIRRAPPGK 240
241 LDLSNSNPTS PVSPSSMASR RGLNIPPTLK QAVSETPFST FSDILDAKSG TLNFKNKAVL 300
301 NSEGVNFSSG SSFRINMSEI IKLEELGKGN YGVVYKALHQ PTGVTMALKE IRLSLEEATF 360
361 NQIIMELDIL HKAVSPYIVD FYGAFFVEGS VFICMEYMDA GSMDKLYAGG IKDEGVLART 420
421 AYAVVQGLKT LKEEHNIIHR DVKPTNVLVN SNGQVKLCDF GVSGNLVASI SKTNIGCQSY 480
481 MAPERIRVGG PTNGVLTYTV QADVWSLGLT ILEMALGAYP YPPESYTSIF AQLSAICDGD 540
541 PPSLPDSFSP EARDFVNKCL NKNPSLRPDY HELANHPWLL KYQNADVDMA SWAKGALKEK 600
601 GEKRS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
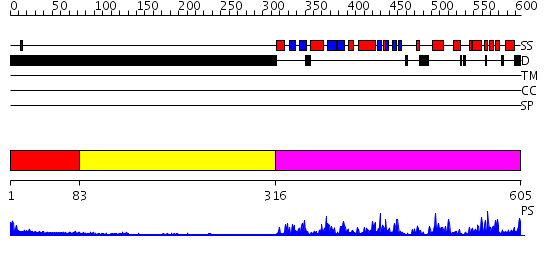
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [83..315] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [316..605] | 48.522879 | No description for 2p55A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)