













| General Information: |
|
| Name(s) found: |
cut14 /
SPBP4H10.06c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1172 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA inner kinetochore of condensed chromosome [IDA cytoplasm [IDA nucleolus [IDA condensin complex [IDA |
| Biological Process: |
mitotic chromosome condensation
[IMP rDNA separation [IMP |
| Molecular Function: |
ATPase activity
[ISS ATP binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKIEELIIDG FKSYAVRTVI SNWDDQFNAI TGLNGSGKSN ILDAICFVLG ITNMSTVRAQ 60
61 NLQDLIYKRG QAGITRASVT IVFNNRDPAS SPIGFENHPQ VSVTRQIIMG GTSKYLINGH 120
121 RALQQNVQNL FQSVQLNINN PNFLIMQGRI TKVLNMKATE ILSMIEEASG TRMFEERKEK 180
181 AFRTMQRKEA KVEEINTLLR EEIEPRLTKL RTEKKTFLEY QHIYNDLERL SHLCTAYDYY 240
241 KLSLKVEELT VQASQKHSHI AEMESSLQTS KQEVLILKEK IKKIEDERMR QMSVSSDRTL 300
301 DSQLQTVNEN ITRISTSIEL KNTALEEEHG DLQQIRGKAK ELETLLRGKR KRLDEVLSVY 360
361 EKRKDEHQSI SKDFKSQEEL ISSLTTGLST TEGHETGYSR KLHEARDTLN DFKAEKETNR 420
421 LKLEGLNKQI SLTKPKKAEA TKRCDQLNRE IDILQNHVEK LKMSLKNTNS DITGEDVLQQ 480
481 KLKQLAKDRG NLLNELDALK SKLAYMEFTY TDPTPNFDRS KVKGLVAQLL TLNEENYDKQ 540
541 TALEITAGGR LYNLIVETEK IGAQLLQKGN LKRRVTIIPL NKITSFVASA ERVGAAKKIS 600
601 NNKAQLALEL IGYDDELLPA MQYVFGSTLV CDTPESAKKV TFHPSVKLKS VTLDGDVYDP 660
661 SGTLTGGSVN KSAGPLLQIQ KLNSLQLKLQ VVTSEYEKLE TQLKDLKTQN ANFHRLEQEI 720
721 QLKQHELTLL IEQRETDSSF RLLSDYQQYK DDVKDLKQRL PELDRLILQS DQAIKKIERD 780
781 MQEWKHNKGS KMAELEKEFN QYKHKLDEFT PILEKSENDY NGVKLECEQL EGELQNHQQS 840
841 LVQGESTTSL IKTEIAELEL SLVNEEHNRK KLTELIEIES AKFSGLNKEI DSLSTSMKTF 900
901 ESEINNGELT IQKLNHEFDR LEREKSVAIT AINHLEKEND WIDGQKQHFG KQGTIFDFHS 960
961 QNMRQCREQL HNLKPRFASM RKAINPKVMD MIDGVEKKEA KLRSMIKTIH RDKKKIQDTV1020
1021 KSIDRFKRSA LEKTWREVNS SFGEIFDELL PGNSAELQPP ENKEFTDGLE IHVKIGSIWK1080
1081 DSLAELSGGQ RSLVALALIM SLLKYKPAPM YILDEIDAAL DLSHTQNIGR LIKTKFKGSQ1140
1141 FIIVSLKEGM FTNANRLFHV RFMDGSSVVQ AR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Liu X, et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
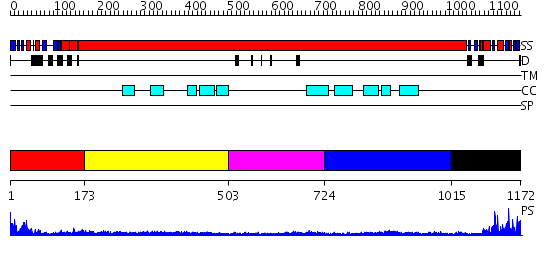
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..172] | 37.69897 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. | |
| 2 | View Details | [173..502] | 2.76 | Tropomyosin | |
| 3 | View Details | [503..723] | 4.73 | Smc hinge domain | |
| 4 | View Details | [724..1014] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1015..1172] | 17.39794 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)