













| General Information: |
|
| Name(s) found: |
slp1 /
SPAC821.08c
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 488 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cell division site [IDA spindle pole body [IDA spindle [IDA condensed nuclear chromosome kinetochore [IDA mitotic checkpoint complex [IDA |
| Biological Process: |
mitotic cell cycle spindle assembly checkpoint
[IGI DNA replication checkpoint [IGI activation of mitotic anaphase-promoting complex activity [TAS mitotic sister chromatid segregation [IGI positive regulation of mitotic metaphase/anaphase transition [IPI cellular protein localization [IGI spindle checkpoint [IPI |
| Molecular Function: |
protein binding
[IPI anaphase-promoting complex binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEIAGNSSTI SPTFSTPTKK RNLVFPNSPI TPLHQQALLG RNGRSSKRCS PKSSFIRNSP 60
61 KIDVVNTDWS IPLCGSPRNK SRPASRSDRF IPSRPNTANA FVNSISSDVP FDYSESVAEA 120
121 CGFDLNTRVL AFKLDAPEAK KPVDLRTQHN RPQRPVVTPA KRRFNTTPER VLDAPGIIDD 180
181 YYLNLLDWSN LNVVAVALER NVYVWNADSG SVSALAETDE STYVASVKWS HDGSFLSVGL 240
241 GNGLVDIYDV ESQTKLRTMA GHQARVGCLS WNRHVLSSGS RSGAIHHHDV RIANHQIGTL 300
301 QGHSSEVCGL AWRSDGLQLA SGGNDNVVQI WDARSSIPKF TKTNHNAAVK AVAWCPWQSN 360
361 LLATGGGTMD KQIHFWNAAT GARVNTVDAG SQVTSLIWSP HSKEIMSTHG FPDNNLSIWS 420
421 YSSSGLTKQV DIPAHDTRVL YSALSPDGRI LSTAASDENL KFWRVYDGDH VKRPIPITKT 480
481 PSSSITIR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
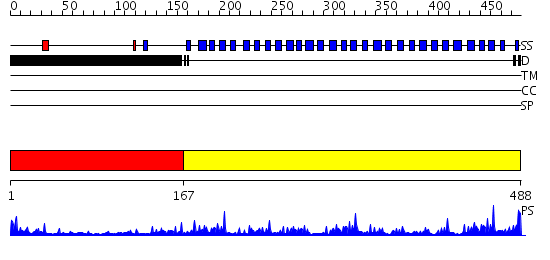
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..166] | 3.221849 | Surface layer protein | |
| 2 | View Details | [167..488] | 57.221849 | PAF-AH Holoenzyme: Lis1/Alfa2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)