













| General Information: |
|
| Name(s) found: |
lon1 /
SPAC22F3.06c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1067 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[ISS mitochondrion [IDA |
| Biological Process: |
misfolded or incompletely synthesized protein catabolic process
[ISS]
|
| Molecular Function: |
ATP binding
[ISS serine-type endopeptidase activity [ISS] ATP-dependent peptidase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MITRLSGACL RRSGAKRNWP REHLVHRSLL ASFSTTQRVL KCSVGPFRTS NVVFKSKEPK 60
61 DNKPLDNKND PKKTHNEDES HTNESLPSTD KKKKKDNDDF KQNLKSSSNK TEEKYSATQA 120
121 SKSKNDEFEL GGEENEDEMP LNGEFNKNVP AKYSVPDVYP QLLALPIARR PLFPGFYKAI 180
181 VTKNPSVSEA IKELIKKRQP YIGAFLLKDE NTDTDVITNI DQVYPVGVFA QITSIFPAKS 240
241 GSEPALTAVL YPHRRIRITE LIPPKEDADS AASSDAAELE TDKSSNLSSN GEVKSDLKQD 300
301 NGKEEPEKEV ESTPSILQNF KVSLVNVENV PNEPFKRQDP VIKAVTSEIM NVFKDIANVS 360
361 PLFREQIANF SISQTSGNVF DEPAKLADFA AAVSAADHRE LQEVLEATNI GDRLQKALYV 420
421 LKKELLNAQL QHKINKEIEQ KITQRHKEYL LTEQLKQIKR ELGQELDSKE ALVTEFKKRT 480
481 ESLSMPDHVK KVFNDELSKF QHLEPMAAEF NITRNYLDWI TQLPWGKRSV ENFDLDHAKE 540
541 VLDRDHYGLK DVKDRVLELV AVGKLRGTMQ GKIMCLVGPP GVGKTSVGKS IASALNREFF 600
601 RFSVGGLTDV AEIKGHRRTY IGAMPGKIVQ ALKKVQTENP LILIDEIDKV GKSHQGDPAS 660
661 ALLELLDSEQ NSAFLDYYMD IPLDVSSVLF VCTANTIDTI PPPLLDRMEV IELSGYVSAE 720
721 KVNIAKGYLI PQAKAACGLK DANVNISDDA IKGLISYYAH ESGVRNLKKS IEKIFRKTSF 780
781 SIVKEIDDEL NSKEKSTGKS GKKTSPQSSE DAANKEASSV PLKVPDKVNI EIEEKDLTKY 840
841 LGPPIYTSQR LYDTTPPGVV MGLGWTPMGG VSMYVETIVK NILSSNSTPS LERTGQLGDV 900
901 MKESSEISYS FSKSFLSKHF PNNKFFEHAR LHMHCPEGSI SKDGPSAGIT MATSLLSLAL 960
961 DTPVPATTAM TGELTLTGKI LRIGGLREKT VAAKLSGMKE ILFPKSNLAD WEQLPDYVKE1020
1021 GLTGVPVAWY DDVFKRVFSN IDAEKCNNLW PNLIKSSSKQ HQISPSH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
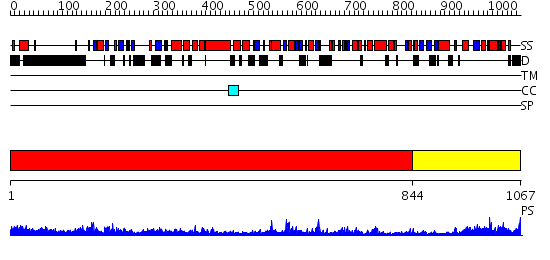
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..843] | 112.0 | Crystal Structure Analysis of ClpB | |
| 2 | View Details | [844..1067] | 47.30103 | Catalytic domain of E.coli Lon protease |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)