













| General Information: |
|
| Name(s) found: |
SPCC1322.05c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 612 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [ISS cytosol [IDA |
| Biological Process: |
leukotriene biosynthetic process
[IEA]
leukotriene metabolic process [IC protein catabolic process [ISS lipid metabolic process [ISS |
| Molecular Function: |
zinc ion binding
[IEA]
leukotriene-A4 hydrolase activity [ISS aminopeptidase activity [ISS epoxide hydrolase activity [ISS metallopeptidase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLRLDPSTQ SNYHDVSISK LDWHARIDFD QELLHGKVSF VIQSARVSQA LSHIILDTSY 60
61 LEIKNVTIND IPTPFRVDKR RGFLGSALHI VPADEIPSSK SCILTILYST TKDCTALQFL 120
121 KPEQTIGGKF PYVFSECQAI HARSFIPCQD TPSVKVPCTF KIRSKLPVIA SGIPCGTANF 180
181 CNGSLEYLFE QKNPIPSYLF CILSGDLAST NIGPRSSVYT EPGNLLACKY EFEHDMENFM 240
241 EAAEQLTLPY CWTRYDFVIL PPSFPYGGME NPNATFATPT LIAGDRSNVN VIAHELAHSW 300
301 SGNLVTNESW QCFWLNEGMT VFLERKILGR LYGEPTRQFE AIIGWGELEE SVKLLGEDSE 360
361 YTKLIQNLEG RDPDDAFSTV PYEKGSNFLY EIERVIGGPS VFEPFLPFYF RKFAKSTVNE 420
421 VKFKHALYEY FSPLGLASKL DSIDWDTWYH APGMPPVKPH FDTTLADPCY KLAESWTNSA 480
481 KNSDDPSKFS SKDIENWSAG QKSLFLDVVY EAVSFPHNYI KRMGDVYSFA ESKNAELSFR 540
541 FFKLALKSKY KPLYNTIAER VGSVGRMKFV RPIFRLLNEA DRAFAIETFE KYKHFYHKIC 600
601 ASQVEKDLGL SE |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
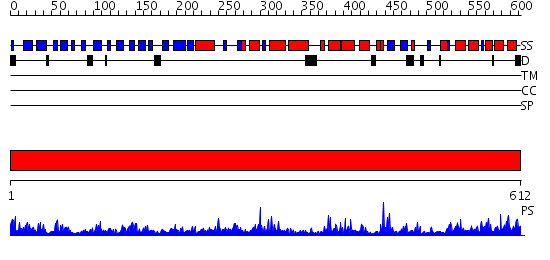
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..612] | 147.0 | Leukotriene A4 hydrolase C-terminal domain; Leukotriene A4 hydrolase N-terminal domain; Leukotriene A4 hydrolase catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)