













| General Information: |
|
| Name(s) found: |
kin1 /
SPBC4F6.06
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 891 amino acids |
Gene Ontology: |
|
| Cellular Component: |
microtubule cytoskeleton
[RCA cytoskeleton [RCA cell tip [IDA cell division site [IDA cytoplasm [IDA |
| Biological Process: |
establishment or maintenance of cell polarity
[IMP cellular response to stress [IEP establishment or maintenance of actin cytoskeleton polarity [IMP protein amino acid phosphorylation [ISS regulation of microtubule-based movement [IMP cytokinesis, site selection [IMP |
| Molecular Function: |
ATP binding
[ISS protein serine/threonine kinase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEYRTNNVPV GNETKSAALN ALPKIKISDS PNRHHNLVDA FMQSPSYSTQ PKSAVEPLGL 60
61 SFSPGYISPS SQSPHHGPVR SPSSRKPLPA SPSRTRDHSL RVPVSGHSYS ADEKPRERRK 120
121 VIGNYVLGKT IGAGSMGKVK VAHHLKTGEQ FAIKIVTRLH PDITKAKAAA SAEATKAAQS 180
181 EKNKEIRTVR EAALSTLLRH PYICEARDVY ITNSHYYMVF EFVDGGQMLD YIISHGKLKE 240
241 KQARKFVRQI GSALSYLHQN SVVHRDLKIE NILISKTGDI KIIDFGLSNL YRRQSRLRTF 300
301 CGSLYFAAPE LLNAQPYIGP EVDVWSFGIV LYVLVCGKVP FDDQNMSALH AKIKKGTVEY 360
361 PSYLSSDCKG LLSRMLVTDP LKRATLEEVL NHPWMIRNYE GPPASFAPER SPITLPLDPE 420
421 IIREMNGFDF GPPEKIVREL TKVISSEAYQ SLAKTGFYSG PNSADKKKSF FEFRIRHAAH 480
481 DIENPILPSL SMNTDIYDAF HPLISIYYLV SERRVYEKGG NWNRIAKTPV SSVPSSPVQP 540
541 TSYNRTLPPM PEVVAYKGDE ESPRVSRNTS LARRKPLPDT ESHSPSPSAT SSIKKNPSSI 600
601 FRRFSSRRKQ NKSSTSTLQI SAPLETSQSP PTPRTKPSHK PPVSYKNKLV TQSAIGRSTS 660
661 VREGRYAGIS SQMDSLNMDS TGPSASNMAN APPSVRNNRV LNPRGASLGH GRMSTSTTNR 720
721 QKQILNETMG NPVDKNSTSP SKSTDKLDPI KPVFLKGLFS VSTTSTKSTE SIQRDLIRVM 780
781 GMLDIEYKEI KGGYACLYKP QGIRTPTKST SVHTRRKPSY GSNSTTDSYG SVPDTVPLDD 840
841 NGESPASNLA FEIYIVKVPI LSLRGVSFHR ISGNSWQYKT LASRILNELK L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
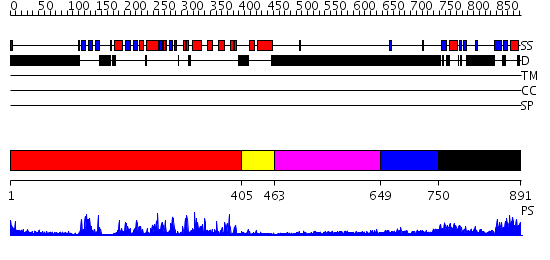
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..404] | 62.522879 | Abl tyrosine kinase, SH3 domain; Abl tyrosine kinase; Abelsone tyrosine kinase (abl) | |
| 2 | View Details | [405..462] | 85.522879 | Catalytic and ubiqutin-associated domains of MARK2/PAR-1: Wild type | |
| 3 | View Details | [463..648] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [649..749] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [750..891] | 39.69897 | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)