













| General Information: |
|
| Name(s) found: |
cmk2 /
SPAC23A1.06c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 504 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell septum
[IDA cytoplasm [IDA |
| Biological Process: |
DNA replication checkpoint
[IMP G2/M transition of mitotic cell cycle [IMP stress-activated protein kinase signaling pathway [IGI negative regulation of cell cycle [IMP cellular response to oxidative stress [IMP protein amino acid phosphorylation [IMP regulation of G2/M transition of mitotic cell cycle [IMP |
| Molecular Function: |
ATP binding
[IEA]
calmodulin binding [TAS protein binding [IPI calcium ion binding [TAS calmodulin-dependent protein kinase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSILAGFKNL LKHSKSSKGR SNASKSVDVS VNRDVAAYTE LAAKNVNAGG DEEIRVANYP 60
61 GLEKYQLIEN LGDGAFSQVY KAYSIDRKEH VAVKVIRKYE MNKKQRQGVF KEVNIMRRVK 120
121 HKNVVNLFDF VETEDFYHLV MELAEGGELF HQIVNFTYFS ENLARHIIIQ VAEAVKHLHD 180
181 VCGIVHRDIK PENLLFQPIE YLPSQNYTPP SLEPNKLDEG MFLEGIGAGG IGRILIADFG 240
241 FSKVVWNSKT ATPCGTVGYA APEIVNDELY SKNVDMWAMG CVLHTMLCGF PPFFDENIKD 300
301 LASKVVNGEF EFLSPWWDDI SDSAKDLITH LLTVDPRERY DIHQFFQHPW IKGESKMPEN 360
361 FTYKPKLHGT PGGPKLSLPR SLVSKGEIDI PTTPIKSATH PLLSSYSEPK TPGVSSVHEA 420
421 MGVAYDIRRL NHLGFSPEQL SKKSMNTGSI KELILDEETT TDDDDYIISS FPLNDTLGSE 480
481 GKDPFSLNLK ESSLYSRRSA KRVN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
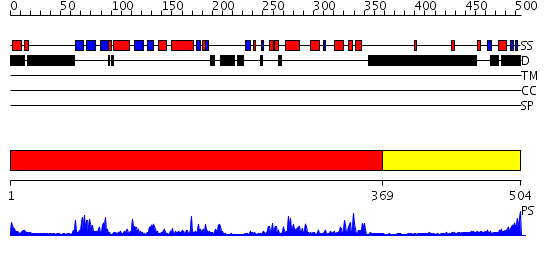
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..368] | 61.30103 | CALMODULIN-DEPENDENT PROTEIN KINASE FROM RAT | |
| 2 | View Details | [369..504] | 9.39794 | Association donain of calcium/calmodulin-dependent protein kinase type II alpha subunit, CAMK2A |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)