













| General Information: |
|
| Name(s) found: |
tif213 /
SPBC17G9.09
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 446 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA cytosol [IDA |
| Biological Process: |
positive regulation of translational initiation
[IC]
translation [IEA] |
| Molecular Function: |
GTP binding
[IEA]
translation initiation factor activity [IEA] GTPase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAENLDISEL SPIHPAIISR QATINIGTIG HVAHGKSTVV KAISGVHTVR FKNELERNIT 60
61 IKLGYANAKI YKCSNEECPR PGCYRSYSSN KEDHPPCEIC NSPMNLVRHV SFVDCPGHDI 120
121 LMATMLNGAA VMDAALLLIA GNESCPQPQT SEHLAAIEIM QLKHIIILQN KVDLIRESAA 180
181 EEHYQSILKF IKGTVAENSP IVPISAQLKY NIDAILEYIV KKIPIPVRDF TTAPRLIVIR 240
241 SFDVNKPGAE VDDLKGGVAG GSILTGVLRL NDEIEIRPGI VTKDDDGRIR CQPIFSRIIS 300
301 LFAEHNDLKI AVPGGLIGVG TTVDPTLCRA DRLVGQVLGS KGNLPEVYTE LEINYFLLRR 360
361 LLGVKSGDKN TTKVQKLAKN EVLMVNIGST STGGRVMMVK ADMAKILLTA PACTEIGEKV 420
421 ALSRRIEKHW RLIGWAKVVE GKTLKV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
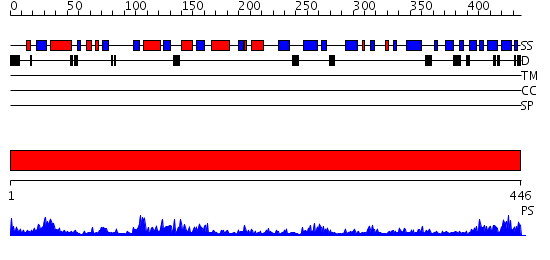
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..446] | 77.69897 | Initiation factor eIF2 gamma subunit, domain II; Initiation factor eIF2 gamma subunit; Initiation factor eIF2 gamma subunit, N-terminal (G) domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)